Abstract
Camellia nitidissima is an endangered species. This species contains two varieties. Here, we report on the chloroplast genomes of C. nitidissima var. nitidissima from Fangcheng (GenBank accession MT157617) and Nanning (MT157618), as well as one sample of C. nitidissima var. microcarpa (MT157619) from Nanning. The total chloroplast genomes of C. nitidissima var. nitidissima Fangcheng and Nanning samples are 156,596 bp and 157,567 bp in length, respectively. C. nitidissima var. microcarpa (MT157619) genome is 157,407 bp in length. The three samples possess GC contents of 37.3%, 128 genes, comprising 86 protein-coding genes, 34 tRNA genes, and 8 rRNA genes.
Camellia nitidissima Chi, belonging to family Theaceae, is distributed across southern China and northern Vietnam (Chang and Ren Citation1998). Camellia nitidissima contains two varieties, C. nitidissima var. nitidisima and C. nitidissima var. microcarpa. It is an ornamental plant and its flowers and leaves can be used as a tea. Because of its high economic value and lack of effective protection, C. nitidissima is seriously exploited. C. nitidissima var. nitidissima is classified as “vulnerable”, and C. nitidissima var. microcarpa as “endangered” (Qin et al. Citation2017). The complete chloroplast (cp) genome of C. nitidissima (GenBank accession NC039645) has been published (Liu et al. Citation2018); however, this study did not consider the disjunctive geographical distribution of C. nitidissima var. nitidissima in Guangxi, China, or high genetic variation of cpDNA sequences from the two disjunctive areas (Lu et al. Citation2020). Here, we present cp genome sequences of C. nitidissima var. nitidissima from Fangcheng (GenBank accession MT157617) and Nanning (MT157618), as well as the cp genome sequence of one Camellia nitidissima var. microcarpa sample (MT157619) from Nanning.
Camellia nitidissima var. nitidissima samples were collected from two disjunctive distribution areas, Nanning Golden Camellia Park and Fangcheng Yellow Camellia National Nature Reserve in Guangxi, China, while C. nitidissima var. microcarpa was collected from Nanning. The Voucher specimens (Liushangli20191215, Liushangli20191216, Liushangli20191231) were deposited in the herbarium of Guangxi Institute of Botany (IBK). GenBank accession numbers for C. nitidissima var. natidisisma are MT157617 and MT157618, and the GenBank accession number for C. nitidissima var. microcarpa is MT157619. Total genomic DNA was extracted from fresh leaves by using the Super Plant Genomic DNA Kit (TIANGEN, Beijing, China). The cp genome was sequenced using an Illumina HiSeq 400 platform. The Filtered reads were assembled with NOVOPlasty (https://github.com/ndierckx/NOVOPlasty). Annotation was performed with the Plastid Genome Annotator (PGA, https://github.com/quxiaojian/PGA).
The cp genome of C. nitidisima var. nitidissima (MT157617) collected from Fangcheng is 156,596 bp in length with 37.3% GC contents, and, consist of a large single-copy (LSC) region (86,272 bp), a small single-copy (SSC) region (18,252 bp), separated by a pair of inverted repeat (IRs: 26,036 bp, each) regions. There are 86 protein-coding genes, 8 rRNA genes, and 34 tRNA genes. The cp genome of C. nitidissima var. nitidissima (MT157618) collected from Nanning is 157,567 bp in length with 37.3% GC contents, and consist of a LSC region (86,632 bp), and, a SSC region separated (17,735 bp) separated by a pair of IRs (26,600 bp) regions. There are 86 protein-coding genes, 8 rRNA genes, and 34 tRNA genes. The cp genome of C. nitidissima var. microcarpa (MT157619) collected from Nanning is 157,047 bp in length with 37.3% GC content, and consist of an LSC region (86,626 bp), an SSC region (18,277 bp), and a pair of IRs: 26,072 bp, each. There are 86 protein-coding genes, 8 rRNA genes, and 34 tRNA genes.
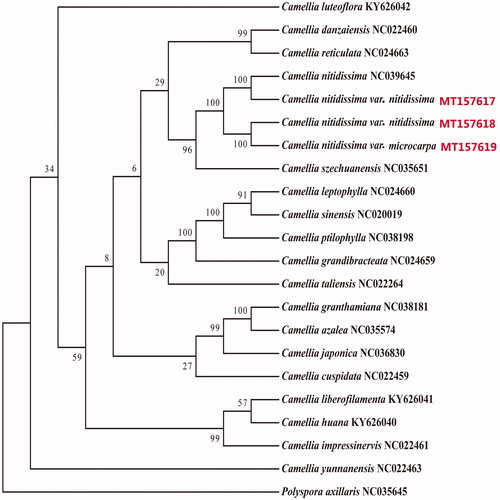
We reconstructed a phylogenetic tree, which was based on 22 complete chloroplast genome sequences, including those of C. nitidissima var. nitidissima (MT157617, MT157618) and C. nitidissima var. microcarpa (MT157619), 18 Camellia species, and the outgroup Polyspora axillaris. Genome sequences were aligned using the software Mafft (Katoh and Standley Citation2013), and the phylogenetic tree was generated by the software RaxML (Stamatakis Citation2014) using maximum-likelihood method with 1000 bootstrap replicates (). The tree shows that C. nitidissima var. nitidissima (MT157618) from Nanning was closely related to C. nitidissima var. microcarpa (MT157619) distributed from Nanning than to the C. nitidissima var. nitidissima sample (MT157619) from Fangcheng (). The complete chloroplast genome of C. nitidissima provides essential data for the conservation and utilization of this species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are available in GenBank, reference number (MT157617, MT157618,MT157619). These data were derived from the following resources available in the public domain: [constructed phylogenetic tree: NC035574, NC038181, NC036830, NC022459, NC022264, NC020019, NC024660, NC038198, NC024659, NC024663, NC022460, NC035651, NC039645, NC022461, KY626040, KY626041, KY626042, NC022463, NC035645, https://www.ncbi.nlm.nih.gov/genbank/]
Additional information
Funding
References
- Chang HT, Ren SX. 1998. Flora reipublicae popularis sinicae. Beijing (China):Science Press; p. 101–112.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu MM, Cao ZP, Zhang J, Zhang DW, Huo XW, Zhang G. 2018. Characterization of the complete chloroplast genome of the Camellia nitidissima, an endangered and medicinally important tree species endemic to Southwest China. Mitochondrial DNA Part B. 3(2):884–885.
- Lu XL, Chen HL, Wei SJ, Bin XY, Ye QQ, Tang SQ. 2020. Chloroplast and nuclear DNA analyses provide insight into the phylogeography and conservation genetics of Camellia nitidissima (Theaceae) in southern Guangxi, China. Tree Genet Genomes. 16:8.
- Qin H, Yang Y, Dong S, He Q, Jia Y, Zhao L, Yu S, Liu H, Liu B, Yan Y, et al. 2017. Threatened species list of China’s higher plants. Biodiv Sci. 25(7):696–744.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.