Abstract
The first complete chloroplast (cp) genome sequences of an endemic and endangered species in China, Calanthe henryi, were reported in this study. The cp genome of C. henryi was 158,256 bp long, with two inverted repeat (IR) regions of 26,348 bp, a large single copy (LSC) region of 87,137 bp, and a small single copy (SSC) region of 18,423 bp. The cp genome of this species contained 113 genes, including 79 protein-coding genes, 4 ribosomal RNA genes, and 30 transfer RNA genes. The overall GC content was 36.7%. Phylogenetic analysis of 60 cp genomes within the subfamily of Epidendroideae suggests that C. henryi is closely related to C. bicolor.
Calanthe henryi Rolfe is a perennial plant in the subfamily of Epidendroideae (Orchidaceae) and usually grows in the evergreen forests from 1,600 m to 2,100 m. It is an endemic species in China and distributed in Hubei, Sichuan (Chen et al. Citation2009), Guizhou (Zhang Citation2007), Hunan (Yu et al. Citation2006) and Jiangxi (Xiao et al. Citation2017) provinces. At present, C. henryi has been regarded as the vulnerable endangered plant by IUCN (China Plant Specialist Group Citation2004). To promote the conservation of this species, we sequenced and analyzed the complete chloroplast (cp) genome of C. henryi using high-throughput sequencing technology.
The fresh leaf of C. henryi was collected from Yushe National Forest Park, Guizhou province, Southwest of China (N26°27′16″, E104°48′8″, 2,205 m). The specimen (lpssy0307) was deposited in the herbarium of the Liupanshui Normal University (LPSNU). Total genome DNA was extracted with CTAB method (Doyle and Doyle Citation1987), which was used for the library construction and sequencing on the Illumina HiSeq 2500 Platform. Approximately 2 Gb raw data were generated and used to de novo assemble the complete cp genome with SPAdes (Bankevich et al. Citation2012). All genes were annotated using PGA (Qu et al. Citation2019) with manual adjustment.
The cp genome of C. henryi (Genbank accession number MT385870) is a typical quadripartite structure with 158,256 bp long, including a pair of inverted repeat (IR, 26,348 bp) regions, a large single copy (LSC, 87,137 bp) region and a small single copy (SSC, 18,423 bp) region. The GC content of the cp DNA is 36.7%, which is similar to the other reported cp genomes from the genus of Calanthe (Dong et al. Citation2018; Miao et al. Citation2019; Zhong et al. Citation2019). A total of 113 unique genes were encoded, including 79 protein-coding (PCD) genes, 30 transfer RNA (tRNA) genes, and 4 ribosomal RNA (rRNA) genes. Of them, 7 PCDs (ndhB, rps12, rpl23, rps7, rps12, rps19 and ycf2), 4 rRNAs (rrn16, rrn23, rrn4.5 and rrn5), and 8 tRNAs (trnA-UGC, trnH-GUG, trnl-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG and trnV-GAC) have two copies. Fifteen genes (atpF, ndhA, ndhB, petB, petD, rpl12, rpl16, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC) contain one intron and three genes (clpP, rps12 and ycf3) have two introns.
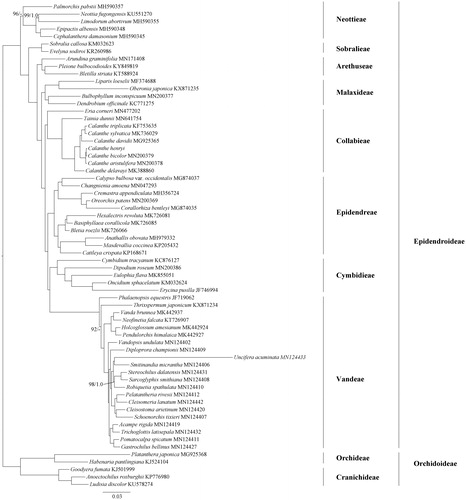
To determine the phylogenetic position of C. henryi within Epidendroideae, the complete cp genome sequences from 59 species of Epidendroideae and five species from Orchidoideae were downloaded from GenBank (). All the complete cp genome sequences were aligned using MAFFT version 7.0 (Katoh and Standley Citation2013). Phylogenomic tree was reconstructed with the maximum likelihood (ML) and Bayesian inference (BI) methods (Ronquist et al. Citation2012; Stamatakis Citation2014). The ML and BI analyses generated the same tree topology (). As shown in the phylogenetic tree (), the species of Calanthe formed one monophyletic clade and C. henryi was close to C. bicolor. The C. henryi cp genome reported in this study may provide useful resources for the development of ornamental and ecological value as well as robust phylogenetic study at deep level of Orchidaceae in the future.
Figure 1. The maximum likelihood (ML) tree of Epidendroideae (Orchidaceae) inferred from the complete chloroplast genome sequences. ML bootstrap percentages (1,000 replicates) and Bayesian inference (BI) posterior probabilities are shown above clades if lower than 100 and 1.0 (dash indicate the brahches that are not supported by posterior probabilities).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov/, reference number MT385870. Submissions are accessioned but the sequence record is not examined and deposited by the GenBank annotation staff before it is free of errors or problems. Therefore, the live link to our dataset is not available at present.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19(5):455–477.
- Chen XQ, Cribb PJ, Gale SW. 2009. Calanthe. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China, Vol. 25. Beijing, St. Louis: Science Press, Missouri Botanic Garden Press. p. 292–301.
- China Plant Specialist Group. 2004. Calanthe henryi. The IUCN Red List of Threatened Species 2004: e.T46641A11073138; [accessed 2020 May 18]. https://dx.doi.org/10.2305/IUCN.UK.2004.RLTS.T46641A11073138.en.
- Dong WL, Wang RN, Zhang NY, Fan WB, Fang MF, Li ZH. 2018. Molecular evolution of chloroplast genomes of orchid species: insights into phylogenetic relationship and adaptive evolution. Int J Mol Sci. 19(3):716.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30(4):772–780.
- Miao LY, Hu C, Huang WC, Jiang K. 2019. Chloroplast genome structure and phylogenetic position of Calanthe sylvatica (Thou.) Lindl. (Orchidaceae). Mitochondrial DNA B. 4(2):2625–2626.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61(3):539–542.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Xiao JW, Xiang XM, Xie D, Wang BQ, Zhang DG, Chen G. 2017. Newly records of medicinal species in Jiangxi. China J Chin Mater Med. 42(22):4431–4435.
- Yu X, Liu K, Huang K, Cao T. 2006. New recorded plants from Hunan province of China. J Cent South Forest Univ. 26(2):143–144.
- Zhang Y. 2007. A list of the orchid plants in Fanjing Mountain National Nature Reserve. Guizhou, China Guizhou Sci. 25(1):43–53.
- Zhong H, Shen L-M, Liu H-P, Liu Z-J, Wu S-S, Zhai J-W. 2019. The complete chloroplast genome of Calanthe arcuata, an endemic terrestrial orchid in China. Mitochondrial DNA B. 4(2):2629–2630.
