Abstract
Hemlock wooly adelgid (HWA), Adelgests ugae Annand (Hemiptera: Adelgidae), is a species native to Asia but later ravages Endangered hemlock forests (Tsuga spp.) in eastern North America. In this study, we obtained the first complete mitochondrial genome of HWA (16,509 bp in length) using meta-genomic sequencing method. The HWA mitogenome has a general gene annotation as other aphids, comprising 13 protein-coding genes, 22 transfer RNAs, and 2 ribosomal RNAs. Our phylogenetic result showed Aphidoidea is sister to Coccoidea and the newly sequenced mitogenome is put on the correct position, sister to Adelgeslaricis.
Hemlock wooly adelgid (HWA), Adelgest sugae Annand, is a species native to Asia, but has caused the collapse of the native hemlock (Tsuga spp.) forest ecosystem in eastern North America (Orwig and Foster Citation1998; Havill et al. Citation2016; Limbu et al. Citation2018). Like other aphid relatives, HWA has host alternation and complex polymorphism in the lifecycles (Sano and Ozaki Citation2012; Limbu et al. Citation2018). In the past, the polymorphism exhibited during the host alternation process makes the identification of aphid species difficult and in recent times, the development of molecular technologies has provided a solution (Stern et al. Citation1997; Foottit et al. Citation2009; von Dohlen Citation2009). HWA is a native aphid species in Taiwan and the secondary host is T. chinensis var. formosana, which is distributed in the high mountains of Taiwan about 2100–3000 meters (Takahashi Citation1937). Morrison spruce, Piceamorrisonicola, is the primary host of HWA in Taiwan, and HWA will induce a small cone-shaped gall on the branch.
Typically, Aphidoidea (belongs to order Hemiptera) divided into three families, Adelgidae, Aphididae, and Phylloxeridae, while HWA belongs to the family Adelgidae (Blackman and Eastop Citation1994, Citation2006; Remaudière and Remaudière Citation1997; Favret et al. Citation2015). In recent years, there have been many reports of complete mitochondrial sequences of aphids, but the species are mainly aphids of the Aphididae (Thao et al. Citation2004; The International Aphid Genomics Consortium, Citation2010; Wang et al. Citation2013, Citation2014; Zhang et al. Citation2014; Wang et al. Citation2015, Citation2016; Ren et al. Citation2016; Zhang, Zheng, et al. Citation2016a; Zhang, Luo Citation2016b; Li et al. Citation2017; Song et al. Citation2019; Wei et al. Citation2019; Zhang et al. Citation2019; Nong et al. Citation2020; Voronova et al. Citation2020). This report has presented the first complete mitochondrial sequence of the Adelgidae.
In May, 2016, the HWA were collected from their hostplant, T. chinensis var. formosanaat the middle-mountainous region of central Taiwan (Meifong, Nantou County, 24°05′15.9″N, 121°10′29.5″E). A colony of the specimens was deposited in the Insect Collections of National Museum of Natural Science (collection number:NMNS ENT 8207-1), Taichung, Taiwan, but five of them were token and extracted their genomic DNAs for next-generation sequencing byMiseq platform. Total 6,403,862 reads (average trimmed length, 213.3 bp) were obtained after removing low DNA quality regions (below Q20) using CLC Genomics Workbench 9 (CLC bio, Aarhus, Denmark). The trimmed reads were de novo assembled into contigs with the setting of 97% sequence similarity via software CLC Genomics Workbench and megahit (Li et al. Citation2015). The mitogenome-like sequences were filtered out by comparing to a Hemiptera reference, which contains 226 related mitogenomic sequences. The assembled contigs were combined and edited to generate mitogenomic sequences using Sequencher 4.10 (GeneCode, Boston, USA). Then, the complete mitogenome of HWA is obtained (Accession number MT263947), 16,059 bp in length. Gene regions and order were predicted using MITOS2 webserver (Bernt et al. Citation2013) and the gene positions were double checked with the public sequences of Adelgeslaricis (KP722589), Diuraphisnoxia (NC_022727), and Diaphorinacitri (NC_030214). The newly sequenced HWA mitogenome shows the same gene order as the reference, Diuraphisnoxia, comprising 13 protein-coding genes, 22 transfer RNAs, and 2 ribosomal RNAs.
To investigate phylogenetic position of HWA, 27 related species were sampled from NCBI to infer Aphid phylogenetic tree (). Maximum likelihood (ML) method (Stamatakis Citation2006; Ott et al. Citation2007) was inferred based on 37 mitochondrial genes. The substitution model was set to GTRGAMMA and the partition scheme was set as gene partition, except for 22 tRNAs, which were concatenated as one partition. Nodal supports were examined using 1000 bootstraps with 10 additional ML searches to improve bootstrapping (Ott et al. Citation2007). The phylogenetic relationship shows that Aleyrodidae is the sister to the clade including Coccoidea and Aphidoidea and the newly sequenced mitogenome, Adelgests nugae, is sister to Adelgeslaricis.
Figure 1. The ML phylogeny of the Aphidoidea, Psylloidea was set as an outgroup and the total aligned length (37 genes) is 15,167 bp.
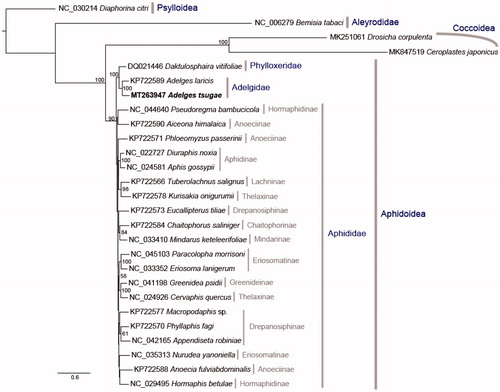
In Virginia, U.S., HWA was first reported in the 1950s and it has now become the most serious pest of native hemlock forests in eastern North America (Limbu et al. Citation2018). It is speculated that the HWA populations of eastern North America are related to the populations from southern Japan based on molecular data (Havill et al. Citation2006, Citation2016). Havill et al. (Citation2016) set the eight monophyletic lineages of HWA based on 748 individuals among 133 sampling sites, but phylogenetic relationships among some of these lineages are still ambiguous. Therefore, our newly produced mitogenome could be used not only to compare with other HWA lineages but also to be a reference to infer phylogenetic relationships within Aphidoidea.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The HWA slide specimens supporting this study were deposited in the Insect Collections of National Museum of Natural Science (collection number:NMNS ENT 8207-1), Taichung, Taiwan, andthe complete HWA mitogenome is available in GenBank (Accession number MT263947; live link: https://www.ncbi.nlm.nih.gov/nuccore/MT263947).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Blackman RL, Eastop VF. 1994. Aphids on the world’s trees: an identification and information guide. Wallingford, UK: CAB International; p. 986.
- Blackman RL, Eastop VF. 2006. Aphids on the world’s herbaceous plants and shrubs. The Atrium, Southern Gate, Chishester, UK: John Wiley & Sons Ltd; p. 1439.
- Favret C, Havill NP, Miller GL, Sano M, Victor B. 2015. Catalog of the adelgids of the world (Hemiptera, Adelgidae). ZK. 534:35–54.
- Foottit RG, Maw HEL, Havill NP, Ahern RG, Montgomery ME. 2009. DNA barcodes to identify species and explore diversity in the Adelgidae (Insecta: Hemiptera: Aphidoidea). Mol Ecol Resour. 9(1):188–195.
- Havill NP, Montgomery ME, Yu G, Shiyake S, Caccone A. 2006. Mitochondrial DNA from hemlock woolly adelgid (Hemiptera:Adelgidae) suggests cryptic speciation and pinpoints the source of the introduction to eastern North America. Ann Entol Soc Am. 99(2):195–203.
- Havill NP, Shiyake S, Galloway A, Foottit R, Yu G, Paradis A, Elkinton J, Montgomery ME, Sano M, Caccone A. 2016. Ancient and modern colonization of North America by hemlock woolly adelgid, Adelges tsugae (Hemiptera: Adelgidae), an invasive insect from East Asia. Mol Ecol. 25(9):2065–2080.
- Li YQ, Chen J, Qiao GX. 2017. Complete mitochondrial genome of the aphid Hormaphis betulae (Mordvilko) (Hemiptera: Aphididae: Hormaphidinae). Mitochondr DNA A DNA Mapp Seq Anal. 28(2):265–266.
- Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 31(10):1674–1676.
- Limbu S, Keena MA, Whitmore MC. 2018. Hemlock woolly adelgid (Hemiptera: Adelgidae): a non-native pest of hemlocks in eastern North America. J Integr Pest Manage. 9(1):27–21.
- Nong X, Wang L, Liu Y, Zhong S, Yu X, Xie Y. 2020. The complete mitochondrial genome of the bamboo aphid Pseudoregmabambucicolaand its phylogenetic position. Mitochondr DNA B. 5(1):642–643.
- Orwig DA, Foster DR. 1998. Forest response to the introduced hemlock woolly adelgid in southern New England, USA. J Torrey Bot Soc. 125(1):60–73.
- Ott M, Zola J, Stamatakis A, Aluru S. 2007. Large-scale maximum likelihood-based phylogenetic analysis on the IBM BlueGene/L. Proceedings of the 2007 ACM/IEEE conference on Supercomputing, Reno; Nevada: ACM.; p. 1–11.
- Remaudière G, Remaudière M. 1997. Catalogue of the World’s Aphididae. Institut National de la RechercheAgronomique, Paris; p. 473.
- Ren ZM, Bai X, Harris AJ, Wen J. 2016. Complete mitochondrial genome of the Rhus gall aphid Schlechtendaliachinensis (Hemiptera: Aphididae: Eriosomatinae). Mitochondr DNA Part B. 1(1):849–850.
- Sano M, Ozaki K. 2012. Variation and evolution of the complex life cycle in Adelgidae (Hemiptera). Entomological Science. 15(1):13–22.
- Song H, Donthu RK, Hall R, Hon L, Weber E, Badger JH, Giordano R. 2019. Description of soybean aphid (Aphis glycines Matsumura) mitochondrial genome and comparative mitogenomics of Aphididae (Hemiptera: Sternorrhyncha)). Insect Biochem Mol Biol. 113:103208.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Stern DL, Aoki S, Kurosu U. 1997. Determining aphid taxonomic affinities and life cycles with molecular data: a case study of the tribe Cerataphidini (Hormaphididae: Aphidoidea: Hemiptera). Syst Entomol . 22(1):81–96.
- Takahashi R. 1937. Phylloxeridae of Formosa (Hemiptera). Trans Nat Hist Soc Formosa, XXVII. 160:11–14.
- Thao ML, Baumann L, Baumann P. 2004. Organization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha). BMC Evol Biol. 4:25.
- The International Aphid Genomics Consortium. 2010. Genome sequence of the pea aphid Acyrthosiphonpisum. PLOS Biol. 8(2):e1000313.
- Von Dohlen CD. 2009. Aphid molecular systematics: history, progress and prospects. Redia. 92:39–45.
- Voronova NV, Levykina S, Warner D, Shulinski R, Bandarenka Y, Zhorov D. 2020. Characteristic and variability of five complete aphid mitochondrial genomes: Aphis fabae mordvilkoi, Aphis craccivora, Myzus persicae, Therioaphis tenera and Appendiseta robiniae (Hemiptera; Sternorrhyncha; Aphididae)). Int J Biol Macromol. 149:187–206.
- Wang Y, Chen J, Jiang LY, Qiao GX. 2015. The complete mitochondrial genome of Mindarus keteleerifoliae (Insecta: Hemiptera: Aphididae) and comparison with other Aphididae insects. Int J Mol Sci. 16(12):30091–30102.
- Wang Y, Huang XL, Qiao GX. 2013. Comparative analysis of mitochondrial genomes of five aphid species (Hemiptera: Aphididae) and phylogenetic implications. PLOS One. 8(10):e77511.
- Wang Y, Huang XL, Qiao GX. 2014. The complete mitochondrial genome of Cervaphis quercus (Insecta: Hemiptera: Aphididae: Greenideinae)). Insect Sci. 21(3):278–290.
- Wang Y, Jiang L, Liu Y, Chen J, Qiao G. 2016. General methods to obtain and analyze the complete mitochondrial genome of aphid species: Eriosomalanigerum (Hemiptera: Aphididae) as an example. Zool Systemat. 41(2):123–132.
- Wei DD, Lang N, Tao Y, He W, Tu YQ, Miao ZQ, Yang L, Wang JJ. 2019. The mitochondrial genome of the brown citrus aphid Aphis (Toxoptera) citricidus: insights into the repeat regions in aphids and phylogenetic implications. Int J Biol Macromol. 136:531–539.
- Zhang B, Ma C, Edwards O, Fuller S, Kang L. 2014. The mitochondrial genome of the Russian wheat aphid Diuraphis noxia: large repetitive sequences between trnE and trnF in aphids. Gene. 533(1):253–260.
- Zhang H, Deng J, Liu Q, Huang X. 2019. The mitochondrial genome of a social aphid, Pseudoregmabambucicola (Hemiptera: Aphididae: Hormaphidinae). Mitochondr DNA Part. 4(2):2100–2101.
- Zhang B, Zheng J, Liang L, Fuller S, Ma CS. 2016a. The complete mitochondrial genome of Sitobion avenae (Hemiptera: Aphididae)). Mitochondr DNA A DNA Mapp Seq Anal. 27(2):945–946.
- Zhang S, Luo J, Wang C, Lv L, Li C, Jiang W, Cui J, Rajput LB. 2016b. Complete mitochondrial genome of Aphis gossypii Glover (Hemiptera: Aphididae)). Mitochondr DNA A DNA Mapp Seq Anal. 27(2):854–855.
