Abstract
Norfolk Robin (Petroica multicolor) is mainly distributed in the islands of the southwest Pacific Ocean. It is an endangered species with a declining population and needs urgent protection. Here, we report the first complete mitochondrial genome of Norfolk Robin in the Petroicidae family. The mitochondrial genome is a typically circular genome with a total length of 16,861 bp, and it contains 13 protein-coding genes, 22 tRNA, and 2 rRNA genes. The GC content is 46.3%, and the percent of A, C, G, and T is 29.9%, 31.8%, 14.5%, and 23.7%, respectively. Phylogenetic analysis based on mitochondrial genomes fully supported that the white-eared catbird is closely related to P. goodenovii. The complete mitochondrial genome obtained from the current study can provide more genomic resources and guidance for the conservation biology, taxonomy and population genetics of Norfolk Robin.
Norfolk Robin (Petroica multicolor: Petroicidae) belongs to the Passeriformes, and its natural habitats are found in the islands of the southwest Pacific Ocean (Kearns et al. Citation2016). Due to continuous threats such as habitat degradation, climate change and predation, and its limited distribution area, its population size has been decreasing. It was evaluated as a distinct endangered species in ICUN (Garnett et al. Citation2011; Kearns et al. Citation2016; BirdLife International Citation2018). Here, we report a Norfolk Robin mitochondrial genome that provides genomic resources for its conservation biology, taxonomy, biodiversity, and population genetics.
The Norfolk Robin were collected from Viti Levu in Fiji (17°29.3′S, 178°06.5′E), and the specimen was deposited at the University of Kansas under the accession no. KU24355. Raw reads of previous study were recovered from the NCBI Sequence Read Archive (SRA) database with the accession number: SRR2968802 (Moyle et al. Citation2016). Trimmomatic v0.33 was used to remove low-quality reads and obtain a total of 5,295,027 high-quality paired-end reads (Bolger et al. Citation2014). After trimming, the CYTBgene (GenBank no.DQ359097) of Norfolk Robin was used as the bait to assemble the mitochondrial genome with the ‘-quick’ mode in MITObim v1.9 software (Hahn et al. Citation2013). The parameter was set to ‘-end 500 –kbait 31’, and iteration was reached to 208 times. The average coverage of the mitochondrial genome is 1481× (247,651 reads), and the final complete mitochondrial genome is determined by the overlapping parts of the mitochondrial genome head and tail. We use the MITOS online annotation website to predict the protein-coding genes (PCGs), tRNAs, and rRNAs of the Norfolk Robin mitochondrial genome (Bernt et al. Citation2013). We further used P. goodenovii as a reference (GenBank no. NC_019667) to manually corrected the start and stop codon of the PCGs according to the standard vertebrate genetic code in Geneious v11.0.2 (Kearse et al. Citation2012). Complete Norfolk Robin mitochondrial genome was stored in the GenBank (GenBank no. MT181966).
Norfolk Robin complete mitochondrial genome is 16,861bp in length, and is annotated with 13 PCGs, 22 tRNAs, 2 rRNAs, and a remnant control region divided by ND6 gene. Its GC content is 46.3%, which is similar to that of most Passeriformes mitochondrial genomes (e.g. 46.4% in Motacilla cinerea). The 13 PCGs were 11,395 bp in length, accounting for 67.6% of the mitochondrial genome. The lengths of the 2 rRNAs, 12S, and 16S rRNA genes were 972 bp and 1594 bp, respectively. The 22 tRNA genes length were 1547, and the non-coding control region was 1291bp in length. Most mitochondrial genes were encoded by the heavy strain, whereas ND6 and eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Pro andtRNA-Glu) were encoded by the light strain. Among the 13 PCGs, ND2 and COX1 used the GTG as the start codon, and other 12 PCGs used ATG as the start codon. Four types of stop codons TAA (ND1, COX2, ATP8, ATP6, ND3, ND4L and CYTB), AGG (COX1), and AGA (ND5), TAG (ND6) were found in the PCGs. In addition, other two genes (COX3 and ND4) and one gene (ND2) terminated with incomplete stop codon T–– and TA–, respectively. They are 5′ terminal of the adjacent gene, which presumptively formed a complete stop codon by post-transcriptional polyadenylation (Anderson et al. Citation1981).
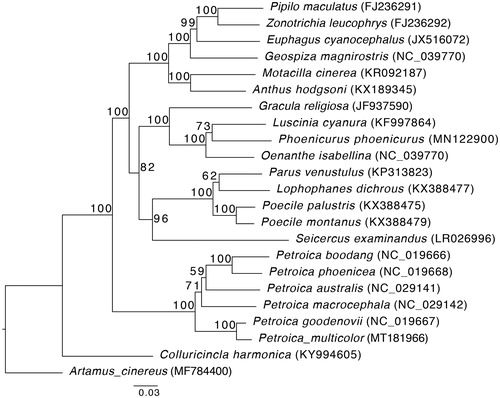
In order to infer the phylogenetic relationship between Norfolk Robin and other Passeriformes, we selected mitochondrial genomes from 21 Passerides species for phylogenetic analysis, with Artamus cinereus (Corvides) set as outgroup (GenBank no. in ). Multiple sequence alignments of mitochondrial genomes of 23Passeriformes species were performed using MAFFTv7.407 (Katoh and Standley Citation2013) with ‘-auto’ option. The maximum likelihood (ML) tree was estimated using IQ-TREE v1.6.7 under the GTR + F + I + G4 model with 1000 ultrafast bootstrapping replicates (Minh et al. Citation2013; Nguyen et al. Citation2015). The ML tree supports Norfolk Robin has the closest relationship with P. goodenovii with full support value (; 100% ML bootstrap), that would contribute to the understanding of the phylogeny of Petroica.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets analyzed during the current study are available from the Sequence Read Archive (SRA) under accession number SRR2968802,https://www.ncbi.nlm.nih.gov/sra/?term=SRR2968802&utm_source=gquery&utm_medium=search. Mitochondrial genome was deposited at NCBI GenBank (MT181966), https://www.ncbi.nlm.nih.gov/nuccore/MT181966.
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290(5806):457–465.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- BirdLife International. 2018. Petroica multicolor (amended version of 2016 assessment). The IUCN Red List of Threatened Species 2018: e.T103734824A129457997.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Garnett S, Szabo J, Dutson G. 2011. The action plan for Australian birds 2010. Melbourne: CSIRO Publishing.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearns AM, Joseph L, White LC, Austin JJ, Baker C, Driskell AC, Malloy JF, Omland KE. 2016. Norfolk Island Robins are a distinct endangered species: ancient DNA unlocks surprising relationships and phenotypic discordance within the Australo-Pacific Robins. Conserv Genet. 17(2):321–335.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Minh BQ, Nguyen MA, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Moyle RG, Oliveros CH, Andersen MJ, Peter AH, Brett WB, Joseph DM, Scott LT, Rafe MB, Brant CF. 2016. Tectonic collision and uplift of Wallacea triggered the global songbird radiation. Nat Commun. 7:12709
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.