Abstract
The sharpsnout seabream Diplodus puntazzo Walbaum, 1792 is a target species of small-scale fishery activities and is cage-cultured for human consumption. Nonetheless, genetic information on this species is limited. We here first sequence its complete mitochondrial genome. The sequence is composed of 16,638 base pairs, accounting for 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, and 2 non-coding regions (D-loop and L-origin). The overall nucleotide composition is: 27.4% A, 28.9% C, 26.9% T, and 16.8% G. Maximum likelihood analyses placed D. puntazzo close to Acanthopagrus and some Pagellus species.
The sharpsnout seabream Diplodus puntazzo Walbaum, 1792 is a coastal and gregarious demersal fish of the family Sparidae, distributed in the eastern Atlantic Ocean and the Mediterranean Sea (e.g. Vinagre et al. Citation2010). It reaches ∼60 cm in length and exceeds 2–3 kilograms in weight (Pavlidis and Mylonas, Citation2011), thus representing a target species of small-scale fishery activities and being cage-cultured for human consumption in the Mediterranean basin (Favaloro et al. Citation2002; Chaouch et al. Citation2013). Notwithstanding its commercial importance, genetic information on this species is still limited to specific mitochondrial regions (COI, cytb, D-loop) (e.g. Bargelloni et al. Citation2005). We fill this gap by sequencing its complete mitochondrial genome (GenBank MT319027). A specimen of D. puntazzo was caught in the Gulf of Pozzuoli (Naples, Tyrrhenian Sea, Mediterranean Sea, 40°48′23.3″N, 14°07′08.1″E), identified based on morphological features by one of us (F. Crocetta) and subsequently deposited in the Darwin Dohrn Museum of the Stazione Zoologica Anton Dohrn of Naples with the code number SZN-OST-0001. Mitochondrial DNA was extracted from dorsal fin tissue (Mascolo, Ceruso, Sordino et al. Citation2019). The assembled and annotated mitogenome was obtained by high-throughput sequencing of enriched mitochondrial DNA with Illumina HiSeq 2500 System (Illumina, San Diego, CA, USA) and bioinformatic analyses were done at Bio-Fab Research (Rome, Italy). The mitogenome is 16,638 base pairs (bp) long, containing 13 protein-coding genes, 2 ribosomal RNA genes (12S and 16S), 22 transfer RNA genes (tRNA), and 2 non-coding regions (D-loop and L-origin). Mitochondrial structure and gene organization are in agreement with typical vertebrate mitogenomes (Wang et al. Citation2008). The majority of mitochondrial genes were encoded on the heavy strand, with the NADH dehydrogenase subunit 6 (ND6) and eight tRNA genes [Gln, Ala, Asn, Cys, Tyr, Ser (UCN), Glu, Pro] being encoded on the light strand. Base composition is 27.4% A, 28.9% C, 26.9% T, and 16.8% G, similar to that observed in other species of the same family (Ceruso, Mascolo, Palma et al. Citation2018; Ceruso, Mascolo, Lowe et al. Citation2018; Mascolo et al. Citation2018a, Citation2018b; Mascolo, Ceruso, Chirollo Citation2019). All protein-coding genes started with an ATG start codon except COI and ND6, that started with GTG and CTA, respectively. Stop codons were of 5 types, i.e. TAA (ND1, ATP6, ND4L), AGG (COI), T (COII, ND3, ND4, ND6, CYTB), TAG (ATP8, ND5), and TA (ND2, COIII). The 12S and 16S rRNA genes were located between the tRNAPhe (GAA) and tRNALeu (TAA) genes and were separated by the tRNAVal gene as in other vertebrates (Li et al. Citation2016). The 22 tRNA genes vary from 66 to 74 bp in length. The control region (968 bp) is located between tRNAPro (TGG) and tRNAPhe (GAA). The non-coding region (L-strand origin of replication) is 36 bp long and is located between tRNAAsn (GTT) and tRNACys (GCA).
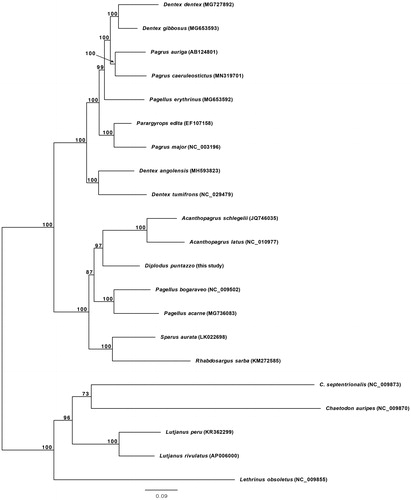
The phylogenetic position of D. puntazzo was investigated performing maximum likelihood (ML) analyses using the RAxML-NG software (Kozlov et al. Citation2019) (). The resultant phylogeny places D. puntazzo close to Acanthopagrus schlegelii and Acanthopagrus latus, with these three species constituting the sister group of Pagellus bogaraveo and Pagellus acarne. The present study informs on phylogenetic relationships within Sparidae and may provide a baseline for future molecular studies on D. puntazzo.
Figure 1. Phylogenetic relationships in the family Sparidae based on the mtDNA sequences available in GenBank and that of Diplodus puntazzo reported here (Acanthopagrus latus NC_010977, Acanthopagrus schlegelii JQ746035, Dentex angolensis MH593823, Dentex dentex MG727892, Dentex gibbosus MG653593, Dentex tumifrons NC_029479, Pagellus acarne MG736083, Pagellus bogaraveo NC_009502, Pagellus erythrinus MG653592, Pagrus auriga AB124801, Pagrus caeruleostictus MN319701, Pagrus major NC_003196, Parargyrops edita EF107158, Rhabdosargus sarba KM272585, Sparus aurata LK022698). Five outgroup species (Chaetodon auripes NC_009870, Chaetodontoplus septentrionalis NC_009873, Lethrinus obsoletus NC_009855, Lutjanus peru KR362299 and Lutjanus rivulatus AP006000) were selected. Maximum likelihood method was used with an automatic bootstrapping cutoff of 0.01.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in Genbank at https://www.ncbi.nlm.nih.gov/nuccore/MT319027, reference accession number MT319027.1.
References
- Bargelloni L, Alarcon JA, Alvarez M, Penzo E, Magoulas A, Palma J, Patarnello T. 2005. The Atlantic-Mediterranean transition: discordant genetic patterns in two seabream species, Diplodus puntazzo (Cetti) and Diplodus sargus (L.). Mol Phylogenet Evol. 36(3):523–535.
- Ceruso M, Mascolo C, Lowe EK, Palma G, Anastasio A, Pepe T, Sordino P. 2018. The complete mitochondrial genome of the common Pandora Pagellus erythrinus (Perciformes: Sparidae). Mitochondr DNA B. 3(2):624–625.
- Ceruso M, Mascolo C, Palma G, Anastasio A, Pepe T, Sordino P. 2018. The complete mitochondrial genome of the common dentex, Dentex dentex (Perciformes: Sparidae). Mitochondr DNA B. 3(1):391–392.
- Chaouch H, Hamida-Ben Abdallah O, Ghorbel M, Jarboui O. 2013. Diet composition and food habits of Diplodus puntazzo (Sparidae) from the Gulf of Gabès (Central Mediterranean). J Mar Biol Ass. 93(8):2257–2264.
- Favaloro E, Lopiano L, Mazzola A. 2002. Rearing of sharpsnout seabream (Diplodus puntazzo, Cetti 1777) in a Mediterranean fish farm: monoculture versus polyculture. Aquac Res. 33(2):137–140.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference . Bioinformatics. 35(21):4453–4455.
- Li J, Yang H, Xie Z, Yang X, Xiao L, Wang X, Li S, Chen M, Zhao H, Zhang Y. 2016. The complete mitochondrial genome of the Rhabdosargus sarba (Perciformes: Sparidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(3):1606–1607.
- Mascolo C, Ceruso M, Palma G, Anastasio A, Sordino P, Pepe T. 2018a. The complete mitochondrial genome of the axillary seabream, Pagellus acarne (Perciformes: Sparidae). Mitochondrial DNA B. 3(1):434–435.
- Mascolo C, Ceruso M, Palma G, Anastasio A, Sordino P, Pepe T. 2018b. The complete mitochondrial genome of the Pink dentex Dentex gibbosus (Perciformes: Sparidae). Mitochondr DNA B. 3(2):525–526.
- Mascolo C, Ceruso M, Chirollo C, Palma G, Anastasio A, Sordino P, Pepe T. 2019. The complete mitochondrial genome of the Angolan dentex Dentex angolensis (Perciformes: Sparidae). Mitochondr DNA B. 4(1):1245–1246.
- Mascolo C, Ceruso M, Sordino P, Palma G, Anastasio A, Pepe T. 2019. Comparison of mitochondrial DNA enrichment and sequencing methods from fish tissue. Food Chem. 294:333–338.
- Pavlidis M, Mylonas C. 2011. Sparidae: biology and aquaculture of Gilthead Sea bream and other species. Wiley-Blackwell, Chichester.
- Vinagre C, Cabral HN, Costa MJ. 2010. Relative importance of estuarine nurseries for species of the genus Diplodus (Sparidae) along the portuguese coast. Estuar Coast Shelf Sci. 86(2):197–202.
- Wang C, Chen Q, Lu G, Xu J, Yang Q, Li S. 2008. Complete mitochondrial genome of the grass carp (Ctenopharyngodon idella, Teleostei): insight into its phylogenic position within Cyprinidae. Gene. 424(1–2):96–101.
