Abstract
Musk Larkspur (Delphinium brunonianum) is a perennial herb of the family Ranunculace with medicinal values. In this study, the chloroplast (cp) genome of this herb was determined to be 153,926 bp long with an A + T-biased base composition, and comprises a pair of inverted repeat (IR) regions (26,559 bp), separated by a large single-copy (LSC) region (84,512 bp) and a small single-copy (SSC) region (16,296 bp). A total of 112 gene species were annotated with 19 of them being completely or partially duplicated. Eighteen gene species harbor one or two introns. Phylogenetic analysis challenged the monophyly of the subfamily Ranunculoideae.
Musk Larkspur (Delphinium brunonianum) is a perennial herb of the family Ranunculace, and is native to southern Tibet of China, Afghanistan, Nepal and northern Pakistan, occurring in grassy or gravelly places with an elevation of 4500–6000 m (Wang and Warnock Citation2001). The dried aerial parts of this herb are used in traditional Tibetan medicine for the treatment of skin itching, various infectious diseases, influenza and snake bites, etc. (Yang Citation1991). Up to now, almost all studies of D. brunonianum have been restricted to its phytochemistry (Asif et al. Citation2019; Zou, Dawa, et al. Citation2019; Zou, Zeren, et al. Citation2019). Little is known about its genetics or genomics, although such a knowledge would be important to its conservation and sustainable exploitation. The present study for the first time presents its complete chloroplast (cp) genome, which is currently accessible from GenBank under the accession number MT457851.
The leaf tissues used for DNA extraction in this study were collected from a single individual of D. brunonianum in Namling County, Shigatse City (89°47′19″E, 29°63′22″N) with the voucher specimen held in (accession number: LQE-2019-067). The high-throughput DNA sequencing was performed on the Illumina HiSeq X Ten Sequencing System (Illumina, CA, USA) by Novogene Biotech Co., Ltd. (Beijing, China). In all, 4.92 M of 150-bp raw reads were used for the assembly of cp genome using MITObim v1.9 (Hahn et al. Citation2013) with that of Delphinium ceratophorum (MK253460) (He et al. Citation2019) as the initial reference. The resultant cp genome sequence was annotated in Geneious Prime 2020 (Biomatters Ltd., Auckland, New Zealand) by comparing with those of its consubfamilial counterparts, e.g., Consolida ajacis (MK569484) (Zhai et al. Citation2019), Delphinium maackianum (MN648402) (Park et al. Citation2020), Delphinium anthriscifolium (MK253461) and Delphinium ceratophorum (MK253460) (He et al. Citation2019).
The cp genome of D. brunonianum is a double-stranded circular molecule of 153,926 bp in size, and comprises a pair of inverted repeat (IR) regions (26,559 bp), separated by a large single-copy (LSC) region (84,512 bp) and a small single-copy (SSC) region (16,296 bp). It has an A + T-biased base composition with an overall A + T content of 61.7% (‘light strand’). A total of 112 gene species were annotated, including 78 protein-coding (PCG), 30 tRNA and four rRNA gene species. Partial or complete gene duplication was detected in 19 gene species, including eight PCGs, seven tRNAs and all four rRNAs. Furthermore, a single intron is present in ten PCG species (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps12 & rps16) and six tRNA gene species (trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA & trnV-UAC), and a couple of introns in two PCG species (clpP & ycf3).
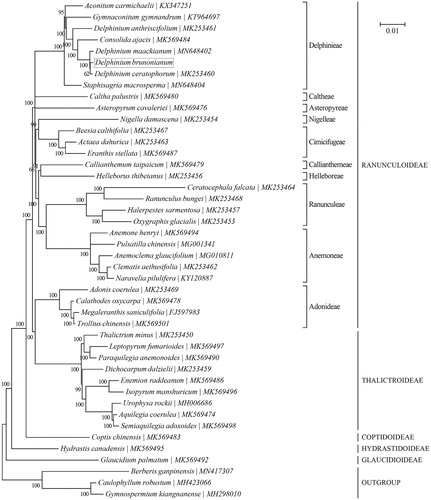
A phylogeny of the family Ranunculaceae was reconstructed based on the Bayesian analysis of concatenated cp protein-coding sequences with MrBayes v3.1.1 (Huelsenbeck and Ronquist Citation2001; Ronquist and Huelsenbeck Citation2003) as in TOPALi v2.5 (Milne et al. Citation2009) (). ‘GTR + G + I’ was selected as the best-fit nucleotide substitution model. The outgroup taxa included in the phylogenetic analysis are three species of the family Berberidaceae, including Berberis ganpinensis (MN417307) (Huang et al. Citation2019), Caulophyllum robustum (MH423066) (Sun et al. Citation2018) and Gymnospermium kiangnanense (MH298010) (Yang et al. Citation2018). The monophyly of the ten tribes within the subfamily Ranunculoideae is well supported by the present study. However, in accordance with previous reports (Wang et al. Citation2009; Cossard et al. Citation2016), our result also challenged the monophyly of Ranunculoideae due to the position of the tribe Adonideae being sister to the subfamily Thalictroideae. In addition, D. brunonianum was found to be most closely related to two of its congeners, i.e., D. ceratophorum (MK253460) and D. maackianum (MN648402).
Figure 1. Phylogenetic relationships of 41 species within the family Ranunculaceae based on the Bayesian analysis of the concatenated coding sequences of chloroplast PCGs. The best-fit nucleotide substitution model is ‘GTR + G+I’. Tribe-level (specifically for the subfamily Ranunculoideae) and subfamily-level taxonomy is presented for each taxon. Three species within the family Berberidaceae were included as outgroup taxa.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT457851.
Additional information
Funding
References
- Asif H, Alamgeer Bukhari IA, Vohra F, Afzal S, Khan SW, Niazi ZR. 2019. Phytochemical analysis of crude extract of Delphinium brunonianum and its effect on hypertension and metabolic perturbations in fructose fed rats. Nat Prod Res. 2:1–5.
- Cossard G, Sannier J, Sauquet H, Damerval C, de Craene LR, Jabbour F, Nadot S. 2016. Subfamilial and tribal relationships of Ranunculaceae: evidence from eight molecular markers. Plant Syst Evol. 302(4):419–431.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucl Acids Res. 41(13):e129–e129.
- He J, Yao M, Lyu R-D, Lin L-L, Liu H-J, Pei L-Y, Yan S-X, Xie L, Cheng J. 2019. Structural variation of the complete chloroplast genome and plastid phylogenomics of the genus Asteropyrum (Ranunculaceae). Sci Rep. 9(1):15285.
- Huang R, Wang C, Liang Q, Wang Y, Yang T-J, Zhang Y. 2019. The complete chloroplast genome of Mahonia eurybracteata subsp. ganpinensis (H.Lév.) T. S. Ying & Boufford (Berberidaceae). Mitochondr DNA Part B. 4(2):3933–3935.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, Wright F. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 25(1):126–127.
- Park S, An B, Park S. 2020. Recurrent gene duplication in the angiosperm tribe Delphinieae (Ranunculaceae) inferred from intracellular gene transfer events and heteroplasmic mutations in the plastid matK gene. Sci Rep. 10(1):2720.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Sun Y, Moore MJ, Landis JB, Lin N, Chen L, Deng T, Zhang J, Meng A, Zhang S, Tojibaev KS, et al. 2018. Plastome phylogenomics of the early-diverging eudicot family Berberidaceae. Mol Phylogenet Evol. 128:203–211.
- Wang W, Lu A-M, Ren Y, Endress ME, Chen Z-D. 2009. Phylogeny and classification of Ranunculales: Evidence from four molecular loci and morphological data. Perspect Plant Ecol Evol Syst. 11(2):81–110.
- Wang W, Warnock MJ. 2001. Delphinium Linnaeus, Sp. Pl. 1: 530. 1753. Flora China. 6:223–274.
- Yang Y-C. 1991. Tibetan medicines. Xining (China): Qinghai People’s Press.
- Yang Z, Peng Z, Zhang H, Lee J, Liu X, Fu C. 2018. The complete chloroplast genome of Gymnospermium kiangnanense (Berberidaceae): an endangered species endemic to Eastern China. Mitochondr DNA Part B. 3(2):713–714.
- Zhai W, Duan X, Zhang R, Guo C, Li L, Xu G, Shan H, Kong H, Ren Y. 2019. Chloroplast genomic data provide new and robust insights into the phylogeny and evolution of the Ranunculaceae. Mol Phylogenet Evol. 135:12–21.
- Zou Y-S, Dawa Z, Lin C-Z, Zhang Q-Y, Yao Y-F, Yuan Y, Zhu C-C, Wang Z-Y. 2019. New amide alkaloids from Delphinium brunonianum. Fitoterapia. 136:104186.
- Zou Y-S, Zeren D-W, Lin C-Z, Yao Y-F, Yuan Y, Zhu C-C. 2019. Chemical constituents from Delphinium brunonianum. J Chin Med Mater. 42(8):1806–1809.
