Abstract
S. miltiorrhiza is a perennial herb of the genus Salvia (Lamiaceae), which is an important medicinal plant for preventing and treating vascular dementia. The complete chloroplast genome sequence of Salvia miltiorrhiza was characterized from Illumina pair-end sequencing. The chloroplast genome of S. miltiorrhiza was 152,680 bp in length, containing a large single-copy region (LSC) of 84,104 bp, a small single-copy region (SSC) of 17,638 bp, and two inverted repeat (IR) regions of 25,469 bp. The overall GC content is 38.70%, while the correponding values of the LSC, SSC, and IR regions are 36.2%, 31.9%, and 43.2%, respectively. The genome contains 131 complete genes, including 86 protein-coding genes (62 protein-coding gene species), 37 tRNA genes (29 tRNA species) and 8 rRNA genes (4 rRNA species). The Neighbour-joining phylogenetic analysis showed that S. miltiorrhiza and Salvia przewalskii clustered together as sisters to other Salvia species.
Introduction
Salvia miltiorrhiza is a perennial herb of the genus Salvia (Lamiaceae), which is an important medicinal plant for preventing and treating vascular dementia and widely distributed in China, which has persisted largely in an undomesticated state that is highly resistant to different environmental stresses. Vascular dementia refers to a disease caused by cerebrovascular disorders, with dementia as the main clinical phase. Modern pharmacological studies have shown that S. miltiorrhiza can significantly increase the number of capillary networks and accelerate blood flow, thereby restoring the function of microcirculation. At the same time, it can also reduce plasma lactic acid content and improve metabolic disorders caused by cell hypoxia. It plays an important role in preventing and treating vascular dementia. Studies have shown that S. miltiorrhiza can reduce the content of lipid peroxide and erythrocyte sorbitol, increase the level of superoxide dismutase, reduce the oxidative stress reaction, and has an antioxidant effect. Studies have also found that S. miltiorrhiza can improve the role of vascular dementia. The mechanism may be: the antioxidant effect of drugs, by restoring the structure and function of nerve cells, inhibiting lipid peroxidation, improving blood rheology, and protecting against ischemia. Damaged neurons improve the pathological response of vascular dementia. S. miltiorrhiza has high ecological and economic value with high levels of intraspecific genetic diversity. S. miltiorrhiza has wide geographic distribution, high intraspecific polymorphism, adaptability to different environments, combined with a relatively small genome size. Consequently, S. miltiorrhiza represents an excellent model for understanding how different evolutionary forces have sculpted the variation patterns in the genome during the process of population differentiation and ecological speciation (Neale and Antoine Citation2011). Moreover, we can develop conservation strategies easily when we understand the genetic information of S. miltiorrhiza. In the present research, we constructed the whole chloroplast genome of S. miltiorrhiza and understood many genome varition information about the species, which will provide beneficial help for population genetics studies of S. miltiorrhiza ().
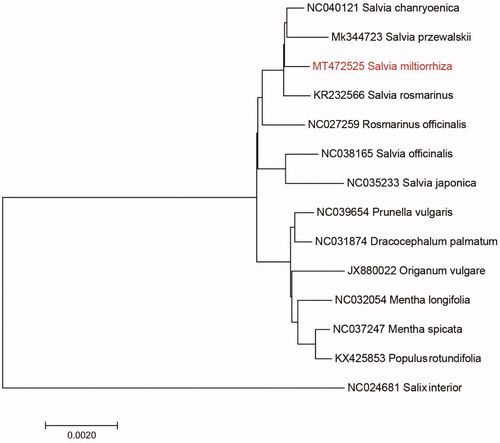
Figure 1. Neighbour-joining (NJ) analysis of S. miltiorrhiza and other related species based on the complete chloroplast genome sequence.
The fresh leaves of S. miltiorrhiza were collected from Chengdu (102°54′E; 30°05′N). Fresh leaves were silica-dried and taken to the laboratory until DNA extraction. The voucher specimen (SDH001) was laid in the Herbarium of Nanyang Institute of Technology and the extracted DNA was stored in the −80 °C refrigerator of the Key Laboratory of School of Biological and Chemical Engineering. We extracted total genomic DNA from 25 mg silica-gel-dried leaf using a modified CTAB method (Doyle Citation1987). The whole-genome sequencing was then conducted by Biodata Biotechnologies Inc. (Hefei, China) with Illumina Hiseq platform. The Illumina HiSeq 2000 platform (Illumina, San Diego, CA) was used to perform the genome sequence. We used the software MITObim 1.8 (Hahn et al. Citation2013) and metaSPAdes (Nurk et al. Citation2017) to assemble chloroplast genomes. We used Salvia przewalskii (GenBank: MK344723) as a reference genome. We annotated the chloroplast genome with the software DOGMA (Wyman et al. Citation2004), and then corrected the results using Geneious 8.0.2 (Campos et al. Citation2016) and Sequin 15.50 (http://www.ncbi.nlm.nih.gov/Sequin/).
The complete chloroplast genome of S. miltiorrhiza (GenBank accession number MT472525) was characterized from Illumina pair-end sequencing. The complete chloroplast genome sequence of Salvia miltiorrhiza was characterized from Illumina pair-end sequencing. The chloroplast genome of S. miltiorrhiza was 152,680 bp in length, containing a large single-copy region (LSC) of 84,104 bp, a small single-copy region (SSC) of 17,638 bp, and two inverted repeat (IR) regions of 25,469 bp. The overall GC content is 38.70%, while the correponding values of the LSC, SSC, and IR regions are 36.2%, 31.9%, and 43.2%, respectively. The genome contains 131 complete genes, including 86 protein-coding genes (62 protein-coding gene species), 37 tRNA genes (29 tRNA species) and 8 rRNA genes (4 rRNA species).
We used the complete chloroplast genomes sequence of S. miltiorrhiza and 13 other related species to construct phylogenetic tree. The 14 chloroplast genome sequences were aligned with MAFFT (Katoh and Standley Citation2013), and then the Neighbour-joining tree was constructed by MEGA 7.0 (Kumar et al. Citation2016). The Neighbour-joining phylogenetic analysis showed that S. miltiorrhiza and Salvia przewalskii clustered together as sisters to other Salvia species.
Disclosure statement
The authors report no conflict of interest.
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov, reference number MT472525.
Additional information
Funding
References
- Campos FS, Kluge M, Franco AC, Giongo A, Valdez FP, Saddi TM, Brito WMED, Roehe PM. 2016. Complete genome sequence of porcine parvovirus 2 recovered from swine sera. Genome Announc. 4(1):01615–e01627.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Neale DB, Antoine K. 2011. Forest tree genomics: growing resources and applications. Nat Rev Genet. 12(2):111–122.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
