Abstract
The complete mitochondrial genome of the double-lined fusileer, Pterocaesio digramma, which belongs to the family Caesionidae was determined. The complete mitochondrial genome has a length of 16,504 bp and consists of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a control region. P. digramma has a mitochondrial gene arrangement that is typical of vertebrates. Phylogenetic analysis using mitochondrial genomes of 15 related species revealed that P. digramma formed a well-supported monophyletic group with the other Caesionidae and Lutjanidae species.
The double-lined fusileer, Pterocaesio digramma (Perciformes, Caesionidae), is a tropical reef-associated marine fish that inhabits the Western Pacific, ranging from Indonesia to Western Australia, New Caledonia, and northwards to Japan. This species is listed as Least Concern in IUCN Red List due to overfishing and harvesting aquatic resources (Russell et al. Citation2016). Although previous studies reported Caesionidae members was nested within the Lutjanidae (Johnson Citation1993; Miller and Cribbs 2007; Guo et al. Citation2014), its taxonomic position within Lutjanidae is not clear. In the present study, we determined the complete mitochondrial DNA sequence of P. digramma and analyzed the phylogenetic relationship of this species with members of Caesionidae and Lutjanidae.
The P. digramma specimen was collected from Ho Chi Minh City, Vietnam (10.53 N, 106.45 W). Total genomic DNA was extracted from the specimen tissue, which has been deposited at the National Marine Biodiversity Institute of Korea (Voucher No. MABIK0002407). The mitogenome was sequenced using Illumina Hiseq 4000 sequencing platform (Illumina, San Diego, CA, USA) and assembled with SOAPdenovo at Macrogen Inc. (Korea). The complete mitochondrial genome was annotated using MacClade ver. 4.08 (http://macclade.org/macclade; Maddison and Maddison Citation2005) and tRNAscan-SE ver. 2.0 (http://lowelab.ucsc.edu/tRNAscan-SE; Lowe and Chan Citation2016).
The complete mitochondrial genome of P. digramma (GenBank accession no. LC549803) is 16,504 bp in length and includes 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a control region. The overall base composition is 27.87% A, 30.87% C, 16.50% G, and 24.76% T. Similar to the mitogenomes of other vertebrates, the AT content is higher than the GC content (Saccone et al. Citation1999). All tRNA genes can fold into a typical cloverleaf structure, with lengths ranging from 67 to 75 bp. The 12S rRNA (949 bp) and 16S rRNA genes (1703 bp) are located between tRNAPhe and tRNAVal and between tRNAVal and tRNALeu(UUR), respectively. Of the 13 protein-coding genes, 12 start with ATG; the exception being COI, which starts with GTG. The stop codon of the protein-coding genes is TAA (COI, ATP8, ND4L, and ND5), T (COII, ND3, ND4, and Cytb), TA (ND2, ATP6, and COIII), and TAG (ND1 and ND6). A control region (825 bp) is located between tRNAPro and tRNAPhe.
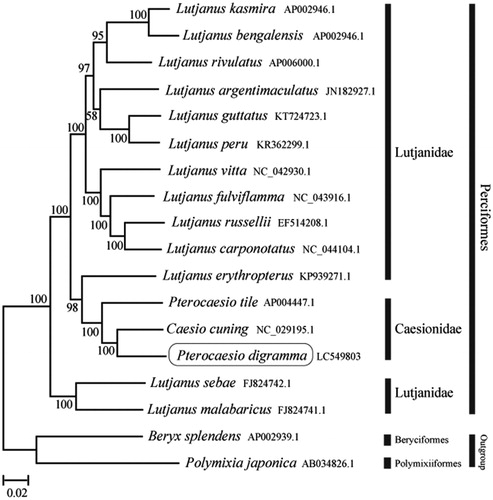
The phylogenetic trees were constructed by the maximum-likelihood method using MEGA 7.0 software (MEGA, Philadelphia, PA, USA; Kumar et al. Citation2016). We compared the phylogenetic trees of the newly sequenced genome and 17 others complete Caesionidae and Lutjanidae species mitochondrial genome sequences acquired from the National Center for Biotechnology Information. We confirmed that P. digramma formed a monophyletic group with the other Caesionidae species and the Caesionidae was nested within the Lutjanidae (). This mitochondrial genome provides a valuable resource for addressing taxonomic issues and developing conservation strategy.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the DNA Data Bank of Japan (accession no. LC549803) at https://www.ddbj.nig.ac.jp.
Additional information
Funding
References
- Guo Y, Bai Q, Yan T, Wang Z, Liu C. 2014. Mitogenomes of genus Pristipomoides, Lutjanus and Pterocaesio confirm Caesionidae nests in Lutjanidae. Mitochondrial DNA A. 27:1–2199.
- Johnson JD. 1993. Percomorph phylogeny: progress and problems. Bull Mar Sci. 52:3–28.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Maddison DR, Maddison WP. 2005. MacClade 4: analysis of phylogeny and character evolution. ver. 4.08. Sunderland (MA): Sinauer Associates.
- Miller TL, Cribb TH. 2007. Phylogenetic relationships of some common Indo-Pacific snappers (Perciformes: Lutjanidae) based on mitochondrial DNA sequences, with comments on the taxonomic position of the Caesioninae. Mol Phylogenet Evol. 44(1):450–460.
- Russell B, Myers R, Smith-Vaniz WF, Carpenter KE, Lawrence A. 2016. Pterocaesio digramma. The IUCN red list of threatened species. e.T20252379A65927523. https://dx.doi.org/10.2305/IUCN.UK.2016-3.RLTS.T20252379A65927523.en.
- Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A. 1999. Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene. 238(1):195–209.