Abstract
Bupleurum falcatum has a long history of use in traditional oriental medicine. The first complete mitochondrial genome sequences of B. falcatum were 463,792 bp based on 494,582 aligned reads. A total of 51 genes was annotated including 32 protein-coding genes, 16 tRNA genes, and three rRNA genes. In a comparison of B. falcatum and carrot (Daucus carota) revealed that the former species has four exclusive genes, but lacks six genes present in the latter. The compositional structure and phylogenetic relationships indicated that the mitochondrial genome of B. falcatum is similar to that of D. carota.
Bupleurum falcatum, a member of family Apiaceae, has a long history of use in traditional oriental medicine. The plant has immunomodulatory, anti-inflammatory, antiviral, anti-tumorigenesis, and anti-ulcer activities (Ashour and Wink Citation2011; Lee et al. Citation2012). A sample was obtained from the National Agrobiodiversity Center (voucher number: IT258576, geographic coordinate: 35°49′19′′N, 127°8′56′′E) in Jeonju, Korea. Whole-genome shotgun sequencing was performed by the Illumina sequencing platform (Illumina, San Diego, CA, USA). First, paired-end (PE) libraries were constructed with an insert size of 270–700 bp, and raw reads with Phred scores ≤30 were removed using the CLC-quality trim tool of CLC Package 4.06 (CLC Inc., Aarhus, Denmark). Second, remaining reads were assembled using the CLC de novo assembly tool, and chloroplast-derived sequences of B. falcatum (Shin et al. Citation2016) were used as a reference. The complete mitochondrion of B. falcatum (Accession no. KX887330) was registered to NCBI GenBank.
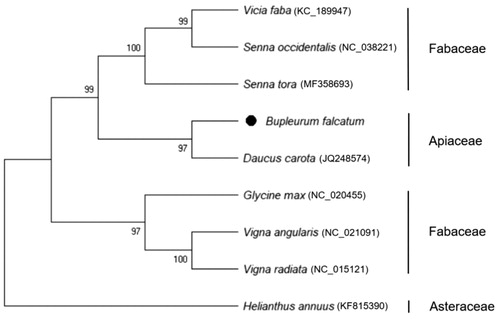
The complete mitochondrial genome of B. falcatum was a circular form of 463,792 bp in length using 494,582 aligned reads with average coverage of 90.5 depth. The genome harbored 51 annotated genes, including 32 protein-coding genes, 16 tRNA genes, and three rRNA genes. The annotated protein-coding genes mostly encode well-known proteins such as electron transport chain (Hofmann Citation2013), rRNAs are commonly used to elucidate evolutionary relationships between organisms (Smit et al. Citation2007), and tRNAs are indispensable components of protein translation (Novoa et al. Citation2012). In a comparison of the same Apiaceae family, B. falcatum and carrot (Daucus carota), their mitochondrial genomes differ significantly in size (463 kb vs. 281 kb) and gene order. However, number of unique genes was similar (51 vs. 53), and total of 47 genes with highly conserved sequences were common to both genomes. Comparison of the mitochondrial genome structures of B. falcatum and D. carota revealed that the former species has four exclusive genes (i.e. rps10, rps14, tatC, and trnX), but lacks six genes (i.e. mttb, rpl2, rpl10, rps7, rps13, and trnfM-CAU) present in the latter. The phylogeney of B. falcatum was reported using genetic markers and ribosomal DNA (Neves and Watson Citation2004); however, mitochondrial phylogeny using the whole-genome sequence in B. falcatum has not yet been reported. Phylogenetic tree was constructed using MEGA X (Kumar et al. Citation2018) with 12 common protein-coding genes (i.e. atp4, atp6, ccmB, ccmC, cob, cox1, cox3, nad2, nad3, nad9, rps3, and rps12) in the 10 reported species. The phylogeny indicated that B. falcatum species is most closely related to the D. carota of Apiaceae family, and B. falcatum is definitely separated from the Fabaceae family (). This study will contribute to develop molecular markers for germplasm classification and breeding of new varieties of Apiaceae.
Geolocation
The genomic DNA sample was obtained from the National Agrobiodiversity Center (voucher number: IT258576, geographic coordinate: 35°49′19′′N,127°8′56′′E) in Jeonju, Korea.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that newly obtained at this study are available in the NCBI under accession number of KX887330 (https://www.ncbi.nlm.nih.gov/nuccore/KX887330.1/).
Additional information
Funding
References
- Ashour ML, Wink M. 2011. Genus Bupleurum: a review of its phytochemistry, pharmacology and modes of action. J Pharm Pharmacol. 63(3):305–321.
- Hofmann NR. 2013. Ribosomal regulation of mitochondrial gene expression. Plant Cell. 25(5):1487.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lee B, Yun HY, Shim I, Lee H, Hahm DH. 2012. Bupleurum falcatum prevents depression and anxiety-like behaviors in rats exposed to repeated restraint stress. J Microbiol Biotechnol. 22(3):422–430.
- Neves SS, Watson MF. 2004. Phylogenetic relationships in Bupleurum (apiaceae) based on nuclear ribosomal DNA its sequence data. Ann Bot. 93(4):379–398.
- Novoa EM, Pavon-Eternod M, Pan T, Ribas de Pouplana L. 2012. A role for tRNA modifications in genome structure and codon usage. Cell. 149(1):202–213.
- Shin DH, Lee JH, Kang SH, Ahn BO, Kim CK. 2016. The complete chloroplast genome of the hare’s ear root, Bupleurum falcatum: its molecular features. Genes. 7(5):20.
- Smit S, Widmann J, Knight R. 2007. Evolutionary rates vary among rRNA structural elements. Nucleic Acids Res. 35(10):3339–3354.