Abstract
The common frogsnail Bufonaria rana, is an ecologically and economically important Tonnoideans in China due to valuable nutrition and pharmacological compounds. However, the taxonomy and phylogeny of the Bursidae have been debated and synonyms among Bursidae species have been reported recently. In this study, we report the first complete mitochondrial genome of Bursidae from B. rana. The mitogenome has 15,510 base pairs (69.0% A + T content) and made up of total of 37 genes (13 protein-coding, 22 transfer RNAs and 2 ribosomal RNAs), and a putative control region. This study was the first available complete mitogenomes of Bursidae and will provide useful genetic information for future phylogenetic and taxonomic classification of Tonnoideans.
Tonnoideans (Tonnoidea) are a moderately diverse super family of Caenogastropoda and it contains about 361 valid species around the world in 21 families (Strong et al. Citation2019), which are the most valuable and vulnerable inshore fisheries resources from subtropical to tropical waters. Bursidae is one of the diverse group in Tonnoidea with 54 recent species, among which several are potentially species complexes (Sanders et al. Citation2017). However, due to limited evaluation of the intraspecific variability and molecular systematic studies, the taxonomy and classification of the Bursidae have been debated and synonyms among species have been reported recently (Ran et al. Citation2020; Strong et al. Citation2019). The common frogsnail Bufonaria rana, widely distributed in the tropical Indo-west Pacific regions, is an ecologically and economically importance species in China which was known as a delicious seafood and a traditional medicine for its pharmacological compounds (Strong et al. Citation2019). The complete mitochondrial genome has been widely used for studying in species identification and classification. Here, we report the first complete mitochondrial genome of Bursidae from B. rana, which will provide a useful genetic markers for species identification and taxonomy assessment.
A tissue samples of B. rana from 5 individuals were collected from HaiNan province, China (Haikou, 20.062794 N, 110.225491 E), and the whole body specimen (#GR0272) were deposited at Marine biological Herbarium, Guangxi Institute of Oceanology, Beihai, China. The total genomic DNA was extracted from the muscle of the specimens using an SQ Tissue DNA Kit (OMEGA, Guangzhou, China) following the manufacturer’s protocol. DNA libraries (350 bp insert) were constructed with the TruSeq NanoTM kit (Illumina, San Diego, CA) and were sequenced (2 × 150bp paired-end) using HiSeq platform at BGI Company, China. Mitogenome assembly was performed by MITObim (Hahn et al. Citation2013). The cytochrome oxidase subunit I (COI) gene of B. granularis (GenBank accession number: MF124183) was chosen as the initial reference sequence for MITObim assembly. Gene annotation was performed by MITOS (http://mitos2.bioinf.uni-leipzig.de).
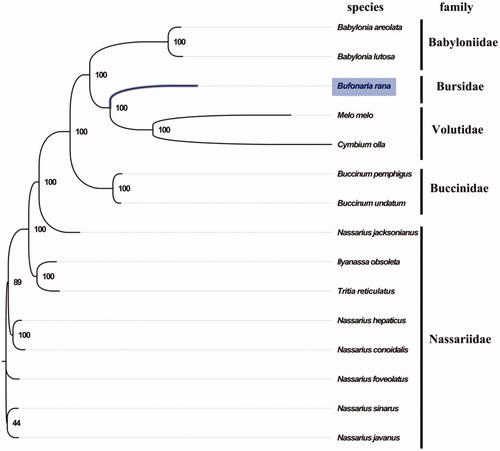
The complete mitogenome of B. rana was 15,510 bp in length (GenBank accession number: MT408027), and containing the typical set of 13 protein-coding, 22 tRNA and 2 rRNA genes, and a putative control region. The overall base composition of the mitogenome was estimated to be A 30.7%, T 38.3%, C 14.9% and G 16.1%, with a high A + T content of 69.0%, which is similar, but higher than Chicoreus asianus (65.2%) (Zhong et al. Citation2020). The mitogenomic phylogenetic analyses showed that B. rana was first clustered with Volutidae then clustered with Babyloniidae (), which is consistent with the phylogenetic analyses of marine gastropod species from Hainan island using COI mitochondrial gene (Ran et al. Citation2020). The complete mitochondrial genome sequence of B. rana was the first sequenced mitogenome in Bursidae, which will be useful for its taxonomy research and future conservation and management.
Figure 1. Phylogenetic tree of 15 species in Subclass Caenogastropoda. The complete mitogenomes is downloaded from GenBank and the phylogenic tree is constructed by maximum-likelihood method with 100 bootstrap replicates. The bootstrap values were labeled at each branch nodes. The gene's accession number for tree construction is listed as follows: Babylonia areolata (NC_023080), Babylonia lutosa (NC_028628), Melo melo (MN462590), Tritia reticulatus (NC_013248), Ilyanassa obsoleta (NC_007781), Nassarius conoidalis (NC_041310), Nassarius hepaticus (NC_038169), Nassarius foveolatus (NC_041546), Nassarius javanus (NC_041547), Nassarius sinarus (NC_041545), Buccinum pemphigus (NC_029373), Buccinum undatum (NC_040940), Nassarius jacksonianus (NC_041548), and Cymbium olla (NC_013245).
Disclosure statement
The authors declare that they have no conflict of interest.
Data availability statement
The data that support the findings of this study are openly available in [National Center for Biotechnology Information] at [https://www.ncbi.nlm.nih.gov/nuccore], reference number [MT408027].
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Ran K, Li Q, Qi L, Li W, Kong L. 2020. DNA barcoding for identification of marine gastropod species from Hainan island, China. Fish Res. 225:105504.
- Sanders MT, Merle D, Bouchet P, Castelin M, Beu AG, Samadi S, Puillandre N. 2017. One for each ocean: revision of the Bursa granularis (Röding, 1798) species complex (Gastropoda: Tonnoidea: Bursidae. J Mollus Stud. 83(4):384–398.
- Strong EE, Puillandre N, Beu AG, Castelin M, Bouchet P. 2019. Frogs and tuns and tritons – a molecular phylogeny and revised family classification of the predatory gastropod superfamily Tonnoidea (Caenogastropoda). Mol Phylogenet Evol. 130:18–34.
- Zhong SP, Huang LH, Liu YH, Huang GQ, Chen XL. 2020. The complete mitochondrial genome of Chicoreus asianus (Neogastropoda: Muricidae) from Beibu Bay. Mitochondrial DNA B. 5(2):1830–1831.
