Abstract
The yellow tip butterfly Anthocharis scolymus (Lepidoptera: Pieridae) has a circular mitochondrial genome of 15,230 bp in size. It consists 13 protein-coding genes, 22 tRNAs, two ribosomal RNA genes, and an AT-rich control region. Using whole mitogenome alignments, we reconstructed the phylogenetic relationships of 28 pierid butterflies. The maximum-likelihood (ML) tree topology was consistent with previous studies.
The Pieridae, one of the largest butterfly families, contains approximately 83 genera and over 1100 species worldwide (Braby Citation2005). This family is currently classified into four monophyletic subfamilies (Dismorphiinae, Pseudopontiinae, Coliadinae, and Pierinae) (Braby and Trueman Citation2006; Wahlberg et al. Citation2014). The genus Anthocharis, which was included within the subfamily Pierinae, consists about 15 species, distributed in North Africa, Eurasia, and North America (Zhou Citation2000; Okumura et al. Citation2016). Recently, the accumulation of whole mitochondrial genomes has facilitated phylogenetic studies in insects (Cameron Citation2014; Liang et al. Citation2018; Zhou et al. Citation2020). Here, we presented the first complete mitogenome of the yellow tip (Anthocharis scolymus), which inhabits mainland China, Far East Russia, the Korean Peninsula, and the Japanese Archipelago in East Asia (Kinoshita Citation1998). Moreover, we present the largest mitogenomic phylogeny of pierid butterflies so far, which would contribute to studies of mitogenomic evolution and phylogeny in butterflies.
An individual male A. scolymus was collected from the campus of Nanjing Forestry University (E118.82, N32.08), Nanjing, Jiangsu Province, China on 16 April 2020 (voucher number: BL_YZ_HJJFD_003, kept in College of Biology and the Environment, Nanjing Forestry University, Nanjing, China). Total genomic DNA was extracted and we followed the major protocol in our previous study (Chen et al. Citation2020) for genomic library construction and sequencing. The generated reads were filtered, trimmed, and mapped to reference mitogenome of A. bambusarum (GenBank: NC_025274) and then assembled using Novoplasty3.8.3 (Dierckxsens et al. Citation2017). The average sequencing coverage is about 800×. We performed automatic annotation using the MITOS web server (Bernt et al. Citation2013) and manually verified the start and stop codons of genes by aligning and comparing with closely related species.
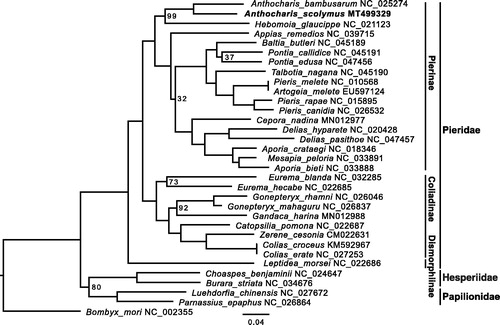
To explore the phylogenetic position of A. scolymus, we collected mitogenomes of 27 additional pierid butterfly species and five outgroups from Genbank. After excluding the control region from the whole mitogenomic alignments, a maximum likelihood (ML) tree was built in RAxML v8.2.10 (Stamatakis Citation2014) with the GTRGAMMA model and 200 ultrafast bootstraps (-f a). The ML tree highly supported (100%) a sister relationship between A. scolymus and A. bambusarum (), and a monophyletic clade of all pierid butterflies. The overall topology is also consistent with previous reports (Espeland et al. Citation2018; Liu et al. Citation2020).
Figure 1. Inferred phylogenetic relationships among pierid butterflies based on whole mitogenome alignment excluding the extremely gappy control region. The domestic silkworm Bombyx mori was used as the outgroup. Bootstrap value at nodes is 100% unless indicated on the tree. GenBank accession numbers of all species used in this study are shown by the species name.
The complete mitochondrial DNA of A. scolymus is a circular molecule of 15,230bp in length with 19.5% GC content. It encodes 37 genes including 13 protein-coding genes, the standard 22 tRNAs, two ribosomal RNA genes, and a putative control region (GenBank accession number: MT499329). The characteristics of the gene order and intergenic spacers in the A. scolymus mitogenome are the same as that of other pierid butterflies (Cao et al. Citation2016).
Disclosure statement
The authors report no conflicts of interests. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT499329.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Braby MF. 2005. Provisional checklist of genera of the Pieridae (Lepidoptera: Papilionoidea). Zootaxa. 832(1):1–16.
- Braby MF, Trueman J. 2006. Evolution of larval host plant associations and adaptive radiation in pierid butterflies. J Evolution Biol. 19(5):1677–1690.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59(1):95–117.
- Cao Y, Hao JS, Sun XY, Zheng B, Yang Q. 2016. Molecular phylogenetic and dating analysis of pierid butterfly species using complete mitochondrial genomes. Genet Mol Res. 15(4):gmr15049196.
- Chen G, Wu H, Wang N, Zhong S, Zhou Y, Liang B. 2020. A mitogenomic phylogeny of spiders and complete mitochondrial genome of Cyriopagopus hainanus (Araneae: Theraphosidae). Mitochondrial DNA B. 5(1):782–783.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Espeland M, Breinholt J, Willmott KR, Warren AD, Vila R, Toussaint EFA, Maunsell SC, Aduse-Poku K, Talavera G, Eastwood R, et al. 2018. A comprehensive and dated phylogenomic analysis of butterflies. Curr Biol. 28(5):770–778.
- Kinoshita M. 1998. Effects of time-dependent intraspecific competition on offspring survival in the butterfly, Anthocharis scolymus (L.) (Lepidoptera: Pieridae). Oecologia. 114(1):31–36.
- Liang B, Wang N, Li N, Kimball RT, Braun EL. 2018. Comparative genomics reveals a burst of homoplasy-free numt insertions. Mol Biol Evol. 35(8):2060–2064.
- Liu G, Chang Z, Chen L, He J, Dong Z, Yang J, Lu S, Zhao R, Wan W, Ma G, et al. 2020. Genome size variation in butterflies (Insecta, Lepidotera, Papilionoidea): a thorough phylogenetic comparison. Syst. Entomol. 45(3):571–582.
- Okumura Y, Ozeki Y, Itoh T, Ohta S, Omura H. 2016. Volatile terpenoids from male wings lacking scent scales in Anthocharis scolymus (Lepidoptera: Pieridae). Appl Entomol Zool. 51(3):385–392.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wahlberg N, Rota J, Braby MF, Pierce NE, Wheat CW. 2014. Revised systematics and higher classification of pierid butterflies (Lepidoptera: Pieridae) based on molecular data. Zool Scr. 43(6):641–650.
- Zhou Y. 2000. Monographia Rhopalocerorum Sinensium. revised ed. Zhengzhou: Henan Science and Technology Press.
- Zhou Y, Wang S, Wang N, Liang Z, Zhong H, Liu Y, Liang B. 2020. Phylogenetic inference of Plebejus argus (Lepidoptera: Lycaenidae) using its complete mitochondrial genome with an extra copy of tRNASer. Mitochondrial DNA B. 5(2):1584–1585.
