Abstract
Japanese Apis cerana honey is believed to ferment due to the presence of some yeast. We analyzed the complete mitochondrial genome of Zygosaccharomyces siamensis, a yeast involved in fermenting honey. Zygosaccharomyces siamensis was obtained from the honey collected from A. cerana hives under traditional beekeeping in the forest of the Minabe-Tanabe Ume system in Wakayama Prefecture, Japan. Its mitochondrial genome was identified as a 23,184 bp circular molecule containing 8 protein-coding genes (PCGs), 24 tRNA genes, and 2 rRNA genes. The PCGs contained a common set of genes encoding ATPase subunits (ATP6, ATP8, and ATP9), three subunits of cytochrome C oxidase (COX1, COX2, and COX3), apocytochrome b (CYTB), and ribosome-associated protein (RPS3). In addition, two open-reading frames coding for LAGLIDADG endonucleases were predicted to be about 1100 bps. The average GC content was found to be 48.6%. The heavy strand was predicted to have 7 PCGs, 22 tRNA genes, and 2 rRNA genes, while the light strand was predicted to contain one PCG and two tRNA genes. Molecular phylogenetic analyses of the mitochondrial DNA genes strongly supported the result obtained from the phylogenetic analysis of partial ITS region sequences, grouping the monophyletic species within the genus Zygosaccharomyces. The complete mitochondrial DNA genome of this honey-fermenting yeast will provide useful information for understanding the basis of the honey fermentation process.
About 4000 years old to the present day, people around the world have been consuming honey collected from honeybees as a valuable carbohydrate source because of its high sugar content (Crane Citation1999). Some types of honey are known to ferment spontaneously, and microorganisms such as yeast, lactic acid bacteria, and acetic acid bacteria have been found in fermented honey (Sinacori et al. Citation2014). In Japan, honey collected from native honeybees kept in the traditional beekeeping style is in a fermented state. Interestingly, honey produced by the European honeybee Apis mellifera bred in Japan does not ferment (Sasaki Citation1999, Citation2001; Yoshida Citation2000, Citation2005). Japanese A. cerana honey is believed to ferment due to the presence of certain yeasts (Sasaki Citation1999; Yoshida Citation2005). The fermentation state of food may change depending on the mitochondrial genes even for the same yeast (Kitagaki and Shimoi Citation2007). Therefore, here, we report the complete mitochondrial genome of the yeast Zygosaccharomyces siamensis, found in honey harvested from a native honeybee hive in Japan (Chikano and Takahasi Citation2019).
We collected raw honey from an A. cerana hive in a forest in the Minabe-Tanabe Ume system, Wakayama Prefecture, Japan (33°49′N 135°24′E), in October 2018. Upon collection, 1 mL of raw honey was immediately transferred to Sabouraud glucose agar medium ReMelt (KANTO KAGAKU, Tokyo, Japan) for mitochondrial DNA analysis. Total DNA was extracted from the yeast colonies grown on the medium using the DNeasy mini kit (QIAGEN, Hilden, Germany) according to the manual accompanying the product. Genomic DNA isolated from yeast in raw honey was sequenced using MiSeq platform (Illumina, San Diego, CA). The complete mitochondrial genome of Zygosaccharomyces mellis (KU920675) was used as a reference sequence (Xiao et al. Citation2017). The resultant reads were assembled and annotated using the MITOS web server (Bernt et al. Citation2013) and Geneious R9 (Kearse et al. Citation2012). Seven of the eight protein-coding gene (PCG) sequences were aligned using MEAG X (Kumar et al. Citation2018). Phylogenetic analysis was performed under the maximum likelihood criterion using TREEFINDER (Jobb Citation2011).
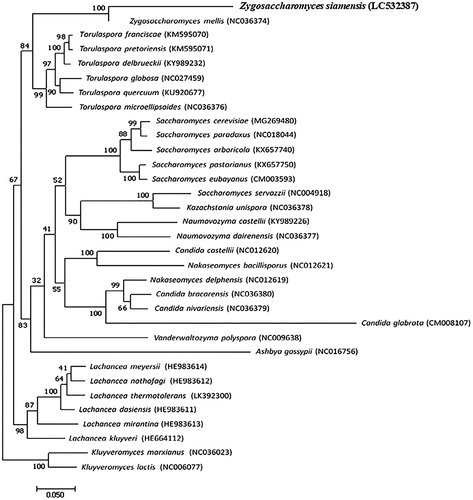
We succeeded in sequencing the entire mitochondrial genome of the yeast Z. siamensis (DDBJ accession number LC532387). These specimens were stored as stocks at the National Museum of Nature and Science, Japan (control number NSMT-I-Hym75320). The genome consisted of a closed loop 23,184 bp long, which included 8 PCGs, 24 tRNA genes, and 2 rRNA genes, which represents a typical yeast mitochondrial genome. The eight PCGs encoded ATPase subunits (ATP6, ATP8, and ATP9), three subunits of cytochrome C oxidase (COX1, COX2, and COX3), apocytochrome b (CYTB), and ribosome-associated protein (RPS3). Two open-reading frames encoding LAGLIDADG endonucleases were predicted to be 1122 and 1143 bp, respectively. Among the eight PCGs, the initiation codon ATG was found in five, ATA in two, and TTA in one gene. Seven PCGs used TAA as the stop codon, whereas one PCG used CAT as the stop codon. Phylogenetic analysis was conducted using seven mitochondrial PCG sequences from 32 closely related taxa (). Zygosaccharomyces siamensis was found to be most closely related to Z. mellis. We expect that the complete sequence data of yeast mitochondrial DNA will provide useful information for understanding the genetic and molecular basis of honey fermentation by yeasts.
Figure 1. Phylogenetic relationships (maximum likelihood) of the yeast species based on the nucleotide sequences of the seven protein-coding genes (COB, COX1, COX2, COX3, ATP6, ATP8, and ATP9) of the mitochondrial genome. The numbers at the nodes indicate the bootstrap support inferred from 1000 bootstrap replicates. Alphanumeric terms indicate the DNA Database of Japan accession numbers.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in DDBJ/GenBank at https://www.ddbj.nig.ac.jp/index.html, accession number DRA009857.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Chikano M, Takahasi J. 2019. Molecular phylogenetic analysis of yeast contained in fermented raw honey. The 40th Annual General Conference. Japanese Society of Food Microbiology. p. 28–29.
- Crane E. 1999. The world history of beekeeping and honey hunting. London: Routledge.
- Jobb G. 2011. Tree finder version of March 2011. Munich. http://www.treefnder.de.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kitagaki H, Shimoi H. 2007. Mitochondrial dynamics of yeast during sake brewing. J Biosci Bioeng. 104(3):227–230.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Sasaki M. 1999. Japanese honeybee-Norther Apis cerana. Tokyo, Japan: Kiayusya.
- Sasaki M. 2001. Science of apiculture. Tokyo, Japan: Science house.
- Sinacori M, Francesca N, Alfonzo A, Cruciata M, Sannino C, Settanni L, Moschetti G. 2014. Cultivable microorganisms associated with honeys of different geographical and botanical origin. Food Microbiol. 38:284–294.
- Xiao S, Nguyen DT, Wu B, Hao W. 2017. Genetic drift and indel mutation in the evolution of yeast mitochondrial genome size. Genome Biol Evol. 9(11):3088–3099.
- Yoshida T. 2000. Rearing method and ecology of Japanese honeybee. Tokyo, Japan: Tamagawa University Press.
- Yoshida T. 2005. Explore the Japanese honeybee society. Tokyo, Japan: Tamagawa University.
