Abstract
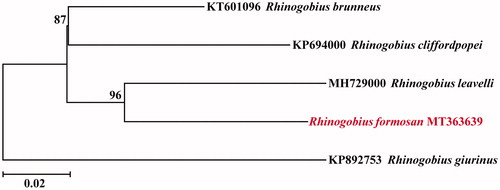
The complete mitochondrial genome of the Rhinogobius formosanus is presented in this study. In brief, it is 16,500 bp long and consists of 13 protein-coding genes, two rRNA genes, 22 tRNA genes, and a control region. The gene order and composition were similar to those of most other vertebrates. The nucleotide compositions of the heavy strand are 16.6% of G, 26.0% of T, 27.7% of A, and 29.8% of C. With the exception of the NADH dehydrogenase subunit 6 (ND6) and eight tRNA genes, all other mitochondrial genes are encoded on the heavy strand. The phylogenetic analysis by neighbour-joining (NJ) method showed that the R. formosanus has the closer relationship to Rhinogobius leavelli in the phylogenetic relationship.
Rhinogobius formosanus Oshima, Citation1919 is previously endemic to river systems of northern and northeastern Taiwan, China (Suzuki et al. Citation2012). However, it is now spreading to other suitable aquatic ecosystem by aquarium trade (Riede Citation2004). Several studies have been carried out regarding the morphology, systematic, migration, and ecology of this species (Oshima Citation1919; Chen and Shao Citation1996; Riede Citation2004; Suzuki et al. Citation2012). However, studies on the genetic diversity of R. formosanus have little been conducted yet. Assessments of genetic information are essential to develop strategies for the identification and management of fisheries resources. The next-generation sequencing (NGS) technologies, such as Illumine, allow considerable numbers of sequence data to be rapidly and efficiently characterized, which makes it particularly feasible for mitogenomes (Gilbert et al. Citation2007). Moreover, Illumine has been successfully used to assemble the mitogenomes of fish species (Cui et al. Citation2009). Therefore, we determined to sequence the complete mitochondrial genome of R. formosanus using the next-generation sequencing (NGS) techniques strategy in order to find DNA markers for the studies on the genetics of R. formosanus.
The specimens of R. formosanus were collected from the ornamental fish market of Sanya (18.11°N, 118.58°E), China during August 2019. All three examined specimens have been deposited in the College of Fisheries and Life Science, Hainan Tropical Ocean University, Sanya, China (Voucher number: HTOU-CFLS-0845 to HTOU-CFLS-0847). The HTOU-CFLS-0845 was used to extract total genomic DNA. The genomic DNA was extracted from dorsal-lateral muscles (30 mg) using Rapid Animal Genomic DNA Isolation Kit (Sangon Biotech Co., Ltd., Shanghai, CN). A genomic library was established followed by next-generation sequencing. Quality check for sequencing data was done by FastQC (Andrews Citation2010) and the fragments sequences were assembled and mapped using Spades v3.9.0 (Bankevich et al. Citation2012).
The final sequence has been deposited in GeneBank with accession number MT363639 (https://www.ncbi.nlm.nih.gov/nuccore/MT363639). The complete mitochondrial genome of R. formosanus (16,500 bp in length) consists of 13 protein-coding genes, 22 transfer RNA genes (tRNA), two ribosomal RNA genes (12S rRNA and 16S rRNA), and two non-coding control regions (control region and origin of light-strand replication). The arrangement of all genes is identical to that of most vertebrates (Wang et al. Citation2008; Chen Citation2013; Chiang et al. Citation2013). Most of the genes are encoded on the heavy strand (H-strand), except for the eight tRNA genes (-Gln, -Ala, -Asn, -Cys,-Tyr, -Ser, -Glu and -Pro) and one protein-coding gene (NADH dehydrogenase subunit 6, ND6). The overall nucleotide compositions of the heavy strand in descending order are 16.6% of G, 26.0% of T, 27.7% of A, and 29.8% of C, with a slight A + T-rich feature (53.7%). All the protein-coding genes begin with an ATG start codon except for COI started with GTG. Three types of stop codons revealed are TAA (COI, ATP8, ATP6, COIII, ND4L, ND5), TAG (ND1, ND2, ND3, ND6), and T (COII, ND4, Cytb). These features are common among vertebrate mitochondrial genome, and TAA is supposed to be appeared via posttranscriptional polyadenylation (Ojala et al. Citation1981). The longest one is ND5 gene (1839 bp) among protein-coding genes, whereas the shortest is ATPase 8 gene (165 bp). The two ribosomal RNA genes, 12S rRNA (951 bp) and 16S rRNA (1684 bp), are located between tRNA-Phe (GAA) and tRNA-Leu (TAA), and are separated by the tRNA-Val gene with the same situation found in other vertebrates. Most genes are either abutted or overlapped. The 22 tRNA genes vary from 69 to 77 bp in length. All these could be folded into the typical cloverleaf secondary structure except tRNA-Ser (AGY), and although numerous non-complementary and T-G base pairs exist in the stem regions. The control region was 842 bp in length, located between tRNA-Pro (TGG) and tRNA-Phe (GAA) gene. The nucleotide composition of control region was 30.17% of A, 21.85% of C, 16.63% of G, 31.35% of T.
To confirm the phylogenetic position of R. formosanus among genus Rhinogobius, a neighbour-joining (NJ) tree was reconstructed with the complete mtDNA sequences from five species. As shown in , the R. formosanus has the closer relationship to Rhinogobius leavelli. The mitogenome information will be beneficial for future phylogenetic studies and specimen identification of Rhinogobius species.
Disclosure statement
The authors declare that they do not have any conflict of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT363639.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. [accessed 2018 Oct 4]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chen IS. 2013. The complete mitochondrial genome of Chinese sucker Myxocyprinus asiaticus (Cypriniformes, Catostomidae). Mitochondrial DNA. 24(6):680–682.
- Chen IS, Shao KT. 1996. A taxonomic review of the Gobiid fish genus Rhinogobius Gill, 1859, from Taiwan, with descriptions of three new species. Zool Stud. 35:200–214.
- Chiang TY, Chen IS, Lin HD, Chang WB, Ju YM. 2013. Complete mitochondrial genome of Sicyopterus japonicus (Perciformes, Gobiidae). Mitochondrial DNA. 24(3):191–193.
- Cui Z, Liu Y, Li CP, You F, Chu HK. 2009. The complete mitochondrial genome of the large yellow croaker, Larimichthys crocea (Perciformes, Sciaenidae): unusual features of its control region and the phylogenetic position of the Sciaenidae. Gene. 432(1–2):33–43.
- Gilbert MTP, Tomsho LP, Rendulic S, Packard M, Drautz DI, Sher A, Tikhonov A, Dalen L, Kuznetsova T, Kosintsev P, et al. 2007. Whole genome shot gun sequencing of mitochondria from ancient hair shafts. Science. 317(5846):1927–1930.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290(5806):470–474.
- Oshima M. 1919. Contributions to the study of the fresh water fishes of the island of Formosa. Ann Carnegie Mus. 12:269–272.
- Riede K. 2004. Global register of migratory species-from global to regional scales. Final Report of the R&D-Project. Bonn (Germany): Federal Agency for Nature Conservation. p. 329.
- Suzuki T, Chen IS, Senou H. 2012. A new species of Rhinogobius Gill, 1859 (Teleostei: Gobiidae) from the Bonin Islands, Japan. J Mar Sci Technol. 19:693–701.
- Wang C, Chen Q, Lu G, Xu J, Yang Q, Li S. 2008. Complete mitochondrial genome of the grass carp (Teleostei, Cyprinidae, Gobioninae). Gene. 424(1–2):96–101.