Abstract
Trapa (Lythraceae) is an aquatic plant genus widely distributed in the old world. Although Trapa species have great edible and medical value, studies related to species identification and utilization are still lacking. Here, we reported the complete chloroplast genome sequence of a cultivated species, T. bicornis. The chloroplast genome size of T. bicornis was 155,539 bp, consisting of a pair of inverted repeat (IR) regions (24,386 bp), separated by a large single copy (LSC) region (88,493 bp) and a small single copy (SSC) region (18,274 bp). A total of 130 genes were annotated, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The phylogenomic analysis supported the monophyly of Trapa, and a sister relationship between T. bicornis and T. natans.
The genus Trapa L. (Myrtales: Lythraceae) comprises approximately 30 species, that are widely distributed in temperate to subtropical regions of Europe, Asia, and Africa (Chen et al. Citation2007). As its fruits are edible and rich in protein and starch, species of this genus have been widely cultivated since the Neolithic (Hoque et al. Citation2009; Artyukhin et al. Citation2019). Today, however, some taxa, e.g. T. muzzanensis and T. verbanensis have become rare and Endangered due to climatic fluctuations, changes in the drainage of many wetlands, ponds and lakes, etc. (Karg Citation2006; Frey et al. Citation2017). Besides, the taxonomy of the genus is extremely confusing worldwide because of the wide variability of morphological traits (Kim et al. Citation2010; Li et al. Citation2017). Thus, more effective molecular markers are needed to foster efforts regarding the identification, conservation, and utilization of Trapa species. Here, we reported the chloroplast genome sequence of T. bicornis Osbeck, which is the first one in cultivated Trapa species, and reconstructed the phylogenetic relationship with other Lythraceae species. Samples of T. bicornis was collected from the vegetable research institute of Suzhou, Jiangsu, China (120.5632°E; 31.3684°N). The voucher specimen was deposited in the Herbarium of Zhejiang University (HZU100718). Genomic DNA was extracted from silica-dried leaf tissue using DNA Plantzol Reagent (Invitrogen, Shanghai, China). DNA library preparation and 125-bp paired-end sequencing were performed on the Illumina HiSeq2500 platform. The chloroplast genome was assembled using NOVOPlasty v.2.63 (Dierckxsens et al. Citation2017), with T. maximowiczii (NC_037023) (Xue et al. Citation2017) as the reference. The resultant genome was annotated in Geneious R11 (http://www.geneious.com) by comparing to T. maximowiczii. The new annotated chloroplast genome sequence was deposited in GenBank (Accession No. MT374084).
The chloroplast genome sequence of T. bicornis was 155,539 bp in length and exhibited the typical quadripartite structure, consisting of a pair of IR regions of 24,386 bp, separated by a LSC region of 88,493 bp and a SSC region of 18,274 bp. The GC contents of the LSC, SSC, and IR regions are 34.2, 30.2, and 42.8%, respectively, with the overall content of 36.4%. The chloroplast genome encoded a total of 130 genes (85 protein-coding, 37 tRNA, and 8 rRNA), of which 18 (7 protein-coding genes, 4 rRNA genes and 7 tRNA genes) were duplicated. Intron-exon structure analysis indicated that 3 protein-coding genes (clpP, ycf3, and rps12) had two introns.
Phylogenetic analyses
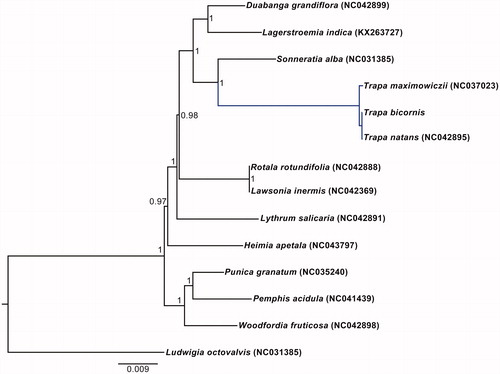
The phylogenetic relationship of Lythraceae was reconstructed using maximum likelihood (ML) method based on the multiple alignment of reported 13 chloroplast genomes within this family, with Ludwigia octovalvis (NC031385) as an outgroup. ML analysis was conducted using PhyML v.3.0 (Guindon et al. Citation2010). The phylogenetic tree strongly supported the sister relationship of Trapa and Sonneratia, which is consistent with previous studies (Berger et al. Citation2016; Yu et al. Citation2018). Within Trapa, T. bicornis was identified as sister to T. natans, and these two species in turn formed a clade sister to T. maximowiczii .
Disclosure statement
The authors are grateful for the open raw genome data in the public database. No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in Genbank with the accession codes MT374084 (https://www.ncbi.nlm.nih.gov/nuccore/MT374084), MT374084.
Additional information
Funding
References
- Artyukhin AE, Mikhaylova EV, Kuluev BR. 2019. Genetic variation of water caltrop (Trapa L.) in several Russian populations. Current challenges in plant genetics, genomics. Bioinf Biotechnol. 24:44.
- Berger BA, Kriebel R, Spalink D, Sytsma KJ. 2016. Divergence times, historical biogeography, and shifts in speciation rates of Myrtales. Mol Phylogenet Evol. 95:116–136.
- Chen JR, Ding BY, Funston AM. 2007. Flora of China. Vol. 13. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 290–291.
- Dierckxsens N, Mardulyn P, Smits G.2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:18.
- Frey D, Reisch C, Narduzzi-Wicht B, Baur E-M, Cornejo C, Alessi M, Schoenenberger N. 2017. Historical museum specimens reveal the loss of genetic and morphological diversity due to local extinctions in the Endangered water chestnut Trapa natans L. (Lythraceae) from the southern Alpine lake area. Botan J Linnean Soc. 185(3):343–358.
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Hoque A, Davey MR, Arima S. 2009. Water chestnut: potential of biotechnology for crop improvement. J New Seeds. 10(3):180–195.
- Karg S. 2006. The water chestnut (Trapa natans L.) as a food resource during the 4th to 1st millennia BC at Lake Federsee, Bad Buchau (Southern Germany). Environ Archaeol. 11(1):125–130.
- Kim C, Ryun Na H, Choi H-K. 2010. Molecular genotyping of Trapa bispinosa and T.Japonica (Trapaceae) based on nuclear AP2 and chloroplast DNA trnL-F region. Am J Bot. 97(12):e149.
- Li X-L, Fan X-R, Chu H-J, Li W, Chen Y-Y. 2017. Genetic delimitation and population structure of three Trapa taxa from the Yangtze River. China Aquat Bot. 136:61–70.
- Xue Z-Q, Xue J-H, Victorovna KM, Ma K-P. 2017. The complete chloroplast DNA sequence of Trapa maximowiczii, Korsh. (Trapaceae), and comparative analysis with other Myrtales species. Aquat Bot. 143:54–62.
- Yu T, Hinsinger DD, Strijk JS, Wee AKS. 2018. The first complete chloroplast genome of a major mangrove species Sonneratia alba Sm. and its implications on conservation efforts. Mitochondrial DNA Part B. 3(2):500–502.