Abstract
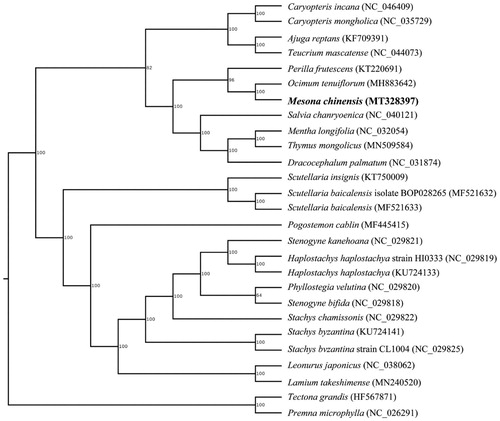
Mesona chinensis is an important traditional Chinese medicine and edible plant resource in China. In this work, we sequenced the complete chloroplast genome of M. chinensis and researched its evolution. The genome size is 152,547 bp, with 37.89% GC content, including a large single copy region (LSC) of 83,482 bp, a small single copy region (SSC) of 17,725 bp and a pair of inverted repeats region (IRs) of 25,670 bp. The complete chloroplast genome was predicted to encode 131 genes, consist of 86 protein-coding genes, 37 tRNA genes and 8 rRNA genes. Phylogenetic analysis showed that M. chinensis was closely related to other Labiatae species Ocimum tenuiflorum.
Chloroplast (cp) is a semi-autonomous organelle in plant cells, which has relatively independent genetic material. Due to its simple structure and highly conserved, cp genome has been widely used in the research of molecular markers, genetic diversity and population variation (Daniell et al. Citation2016). In recent years, cp genomes of many medicinal plants have been reported, which will helpful to develop DNA barcode and improve the understanding of genetic structure (Qian et al. Citation2013; Zhang et al. Citation2016; Shen et al. Citation2017). However, the cp genome of M. chinensis is still unclear.
Mesona chinensis, belonging to the Labiatae family, is an importante conomic and medicinal plants, widely distroibuted in Fujian, Guangdong and Guangxi provinces, has the effects of antioxidation and hypolipidemic (Lin et al. Citation2017). In this study, the structural characteristics of cp genome of M. chinensis were described, which provides an important basis for further study. Firstly, fresh leaves of M. chinensis were collected from Guangdong Academy of Agricultural Sciences (Guangzhou, China). Then, the genomic DNA was extracted using plant genomic DNA kit (Omega), which was strored at Key Laboratory for Crops Genetic Improvement of Guangdong in Guangdong Academy of Agricultural Sciences (specimen code Lfc2019) and seuquenced on the Novaseq platform (Illumina, San Diego, CA). Finally, we assembled and annotated the cp genome by GetOrganelle and Geseq, respectively (Tillich et al. Citation2017; Jin et al. Citation2019).
The size of the cp genome was 152,547 bp, with 37.89% GC content, exhibited a typical quadripartite structure, consisted of a pair of IR regions (25,670 bp) separated by LSC region of 83,482 bp and SSC region of 17,725 bp (GenBank assession number MT328397.1). Total of 131 functional genes were predicted, including 86 protein-coding genes, 37 tRNA genes and 8 rRNA genes. The phylogenetic relationship of M. chinensis was constructed with the complete chloroplast genomes of other 26 species using maximum likelihood method by RaxML software (Stamatakis Citation2014). As shown in , the position of M. chinensis was closely to Ocimum tenuiflorum with a bootstrap of 100%. The characterized cp genome sequence of M. chinensis will be facilitate future research on molecular breeding and genetic engineering.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in [GenBank] at [https://www.ncbi.nlm.nih.gov/nuccore/MT328397.1], reference number [MT328397.1].
Additional information
Funding
References
- Daniell H, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):134.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2019. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv, 256479. https://doi.org/10.1101/256479
- Lin L, Xie J, Liu S, Shen M, Tang W, Xie M. 2017. Polysaccharide from Mesona chinensis: extraction optimization, physicochemical characterizations and antioxidant activities. Int J Biol Macromol. 99:665–673.
- Qian J, Song J, Gao H, Zhu Y, Xu J, Pang X, Yao H, Sun C, Li XE, Li CY, et al. 2013. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS One. 8(2):e57607.
- Shen XF, Wu ML, Liao BS, Liu ZX, Bai R, Xiao SM, Li XW, Zhang BL, Xu J, Chen SL. 2017. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules. 22(8):1330.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zhang R, Li Q, Gao J, Qu M, Ding P. 2016. The complete chloroplast genome sequence of the medicinal plant Morinda officinalis (Rubiaceae), an endemic to China. Mitochondrial DNA Part A. 27(6):4324–4325.