Abstract
Monochamus alternatus alternatus is the major vector of pinewood nematode, Bursaphelenchus xylophilus, in Asia. The length of the complete mitochondria genome of M. alternatus alternatus was 15,880 bp with 21% GC content, including 39.7% A, 12.3% C, 8.7% G and 39.3% T. There were 13 protein-coding genes, 22 tRNAs, 2 rRNAs, and one AT-rich region. This study provides a useful genetic information for subsequent study of the differences between M. alternatus subspecies.
Monochamus alternatus is one of the trunk borer of pine trees and is also the main transmission vector of pinewood nematode that is the agent of the pine wilt disease (Nickle et al. Citation1981). The study have divided the beetle into two subspecies, including Monochamus alternatus alternatus Hope from China and Vietnam, and Monochamus alternatus endai Makihara from Japan and South Korea (Makihara Citation2004). However, because the relevant genomic analysis of each subspecies is not detailed enough at present, the relationship between them in genetic evolution in not clear. We determined the complete mitochondria genome of Monochamus alternatus alternatus that from Vietnam in this study. The results could provide an important genetic information to study the genetic evolution of M. alternatus subspecies.
The M. alternatus alternatus adults were collected from Dingli county, Liangshan province, Vietnam (107°03′50″E, 21°35′54″N) by the traps with sexual attractants. The specimens were stored in the Fujian Agriculture and Forestry University (MA-201912). The total DNA was extracted by Insect DNA Kit (Omega Bio-Tek, GA, USA) and sequenced by the Illumina Hiseq 2500 (Illumina, CA, USA) at the Novogene (Beijing, China). About 48,469,116 reads were assembled by GetOrganelle (Jin et al. Citation2020). The quality of mitogenome assembly showed that the customized error rate was 0.0059 ± 0.0086. Then, the mitogenome was annotated by GeSeq (Tillich et al. Citation2017), and manually adjusted in Geneious (Kearse et al. Citation2012) according to M. alternatus (Genbank Accession No. KJ809086.1). The complete mitogenome sequence of M. alternatus alternatus has been submitted to NCBI Genbank with accession number MT547196.
The complete mitochondria genome length of M. alternatus alternatus was 15,880 bp with 39.7% A, 39.3% T, 12.3% C, and 8.7% G, and the GC content was 21%. The complete mitochondria genome encoded 13 protein-coding genes, 22 tRNAs, 2 rRNAs, and one AT-rich region. The content and order of these genes in the mitochondrial genome of M. alternatus were consistent with those of other coleopteran insects (Kim et al. Citation2009). The complete mitochondrial genome of Japanese pine sawyer, Monochamus alternatus have been analyzed, and the content and order of code genes were same between Japanese pine sawyer and M. alternatus alternatus alternatus in this study (Li et al. Citation2016).
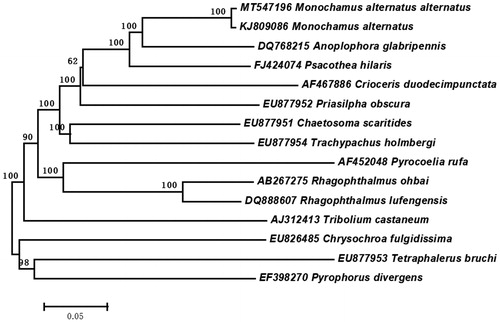
According to the mitochondrial genome sequence of M. alternatus alternatus, the sequence alignment was performed with the MAFFT (Katoh and Standley Citation2013). The phylogenetic analysis was constructed with 15 different species of coleoptera by MEGA 6.0 using neighbor-joining tree model with 1000 bootstrap replicates. The result showed that the M. alternatus alternatus and Japanese pine sawyer were clustered together, and sister to Anoplophora glabripennis (). The complete mitochondrial genome of M. alternatus alternatus will provide useful genetic information to study the genetic evolution of M. alternatus subspecies, as well as in insects of Monochamus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT547196.
Additional information
Funding
References
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol 21, 241. https://doi.org/10.1186/s13059-020-02154-5
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kim KG, Hong MY, Kim MJ, Im HH, Kim MI, Bae CH, Seo SJ, Lee SH, Kim I. 2009. Complete mitochondrial genome sequence of the yellow-spotted long-horned beetle Psacothea hilaris (Coleoptera: Cerambycidae) and phylogenetic analysis among coleopteran insects. Mol Cells. 27(4):429–441.
- Li F, Zhang H, Wang W, Weng H, Meng Z. 2016. Complete mitochondrial genome of the Japanese pine sawyer, Monochamus alternatus (Coleoptera: Cerambycidae). Mitochondr DNA. 27(2):1144–1145.
- Makihara H. 2004. Two new species and a new subspecies of Japanese Cerambycidae (Coleoptera). Bull Forestry Forest Prod Res Inst. 3:15–24.
- Nickle WR, Golden AM, Mamiya Y, Wergin WP. 1981. On the taxonomy and morphology of the pine wood nematode, Bursaphelenchus xylophilus (Steiner & Buhrer 1934) Nickle. J. Nematol. 13:385–392.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:6–11.