Abstract
The first complete chloroplast genome (cpDNA) sequence of Taxus yunnanensis was determined from Illumina HiSeq pair-end sequencing data in this study. The cpDNA is 129,190 bp in length. Like other species of taxus genus, the chloroplast genome of T. yunnanensis has lost one of the large inverted repeats (IRs). The genome contains 116 genes, including 82 protein-coding genes, 4 ribosomal RNA genes, and 30 transfer RNA genes. Further phylogenomic analysis showed that T. yunnanensis closed to T. brevifolia in Lauraceae family.
Taxus yunnanensis is the species of the genus Taxus within the family Taxaceae, an evergreen tree commonly knwon as Hongdoushan, and grown mainly in Yunnan Province of China (Banskota et al. Citation2003; Ha et al. Citation2014). More than 550 taxane diterpenoids have been isolated from Taxus species (Wang et al. Citation2011). Taxus yunnanensis is considered as a promising source of taxane diterpenes, and more Taxol derivatives have biological activities such as antitumor, anticancer, anti-inflammatory, antiviral (Baloglu and Kingston Citation1999; Wang et al. Citation2011). Therefore, T. yunnanensis has huge medicinal value. However, there has been no genomic studies on T. yunnanensis.
Herein, we reported and characterized the complete T. yunnanensis plastid genome. The GenBank accession number is MT536348. One T. yunnanensis individual (specimen number: 2020013) was collected from Kunming, Yunnan Province of China (25°14 '28" N, 102°75 '43" E). The specimen is stored at Yunnan Academy of Forestry Herbarium, Kunming, China and the accession number is ZJFEP223. DNA was extracted from its fresh leaves using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA, USA).
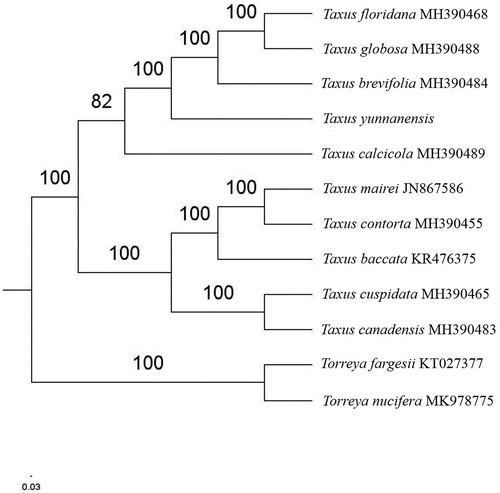
Paired-end reads were sequenced by using Illumina HiSeq system (Illumina, San Diego, CA). In total, about 20.2 million high-quality clean reads were generated with adaptors trimmed. Aligning, assembly, and annotation were conducted by CLC de novo assembler (CLC Bio, Aarhus, Denmark), BLAST, GeSeq (Tillich et al. Citation2017), and GENEIOUS v 11.0.5 (Biomatters Ltd, Auckland, New Zealand). To confirm the phylogenetic position of T. yunnanensis, other thirteen species of Taxus genus from NCBI were aligned using MAFFT v.7 (Katoh and Standley Citation2013). The Auto algorithm in the MAFFT alignment software was used to align the twelve complete genome sequences and the G-INS-i algorithm was used to align the partial complex sequences. The maximum likelihood (ML) bootstrap analysis was conducted using RAxML (Stamatakis Citation2006); bootstrap probability values were calculated from 1000 replicates. Torreya fargesii (KT027377) and Torreya nucifera (MK978775) were served as the out-group.
The complete T. yunnanensis plastid genome is a circular DNA molecule with the length of 129,190 bp, the overall GC content of the whole genome is 34.6%, like other species of taxus genus, the chloroplast genome of T. yunnanensis has lost one of the large inverted repeats (IRs). The plastid genome contained 116 genes, including 82 protein-coding genes, 4 ribosomal RNA genes, and 30 transfer RNA genes. Phylogenetic analysis showed that T. yunnanensis closed to T. brevifolia in Taxus genus (). The determination of the complete plastid genome sequences provided new molecular data to illuminate the Taxus genus evolution.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in NCBI GenBank database at (https://www.ncbi.nlm.nih.gov) with the accession number is MT536348, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Additional information
Funding
References
- Baloglu E, Kingston D. 1999. The taxane diterpenoids. J Nat Prod. 62(10):1448–1472.
- Banskota AH, Tezuka Y, Nguyen NT, Awale S, Nobukawa T, Kadota S. 2003. DPPH radical scavenging and nitric oxide inhibitory activities of the constituents from the wood of Taxus yunnanensis. Planta Med. 69(6):500–505.
- Ha P, Wen SZ, Li Y, Gao Y, Jiang XJ, Wang F. 2014. New taxane diterpenoids from Taxus yunnanensis. Nat Prod Bioprospect. 4(1):47–51.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang YF, Shi QW, Dong M, Kiyota H, Gu YC, Cong B. 2011. Natural taxanes: developments since 1828. Chem Rev. 111(12):7652–7709.