Abstract
The genus Ptychoptera Meigen, Citation1803 is the largest genus of the family Ptychopteridae with 78 known species. In this study, we report a nearly complete mitochondrial (mt) genome of this genus, which is a circular molecule of more than 15,028 bp. The mt genome contains 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes and a non-coding region. The overall base composition is A (38.1%), T (36.7%), C (14.9%), and G (10.4%), with an AT content of 74.8%. The AT content of N-strand genes (75.7%) is higher than that of the J-strand genes (71.7%). Phylogenetic analysis reveals that the monophyly of Ptychopteridae, Bibiomorpha, Tipulomorpha and Brachycera are strongly supported, and the sister group relationship between Tanyderidae and Ptychopteridae is not supported.
The family Ptychopteridae includes three genera, Bittacomorpha Westwood, Citation1835, Bittacomorphella Alexander, Citation1916, and Ptychoptera Meigen, Citation1803, of which Ptychoptera is the largest genus and widely distributed in the world except for the Australasian (Oceanian) region with 78 described species (Kang et al. Citation2013a, Citation2013b; Paramonov Citation2013Citation; Török et al. 2015). More mitochondrial (mt) genomes have been widely used for reconstructing phylogenetic relationships in many insect groups, including the lower Diptera (Beckenbach Citation2012; Timmermans and Vogler Citation2012; Wang et al. Citation2012; Caravas and Friedrich Citation2013; Li et al. Citation2013; Li et al. Citation2015; Wang et al. Citation2017). There are nearly 700 complete or nearly complete lower dipteran mt genomes available in GenBank, of which only two mt genomes are from Ptychopteridae. Here, we report another nearly complete mt genome of Ptychopteridae.
The specimen of Ptychoptera qinggouensis Kang, Yao and Yang, 2013 used in this study was collected from Daqinggou, Kezuohouqi, Neimenggu, China (42°45′47ʺN 122°12′14ʺE, 200 m) and stored in the Entomological Museum of Qingdao Agricultural University, China (No. PTY0001). Qualified DNA samples were pooled for next-generation sequencing library construction following the method proposed by Gillett et al. (Citation2014). The standard PCR reactions were sequenced by primers designed by Simon et al. (Citation2006). BLAST searches were conducted with BioEdit 7.0.5.3 for the bait sequence against mt genome assemblies. The sequence was annotated following the method proposed by Cameron (Citation2014). Maximum-likelihood analysis was conducted by RAxML v7.0.3 (Stamatakis Citation2006).
The nearly complete mt genome of P. qinggouensis (GenBank accession no. MT380468) is 15,028 bp in length. It contains 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes and a non-coding region. The overall base composition is A (38.1%), T (36.7%), C (14.9%), and G (10.4%), with an AT content of 74.8%. It has a weakly positive AT-skew and a negative GC-skew. For protein-coding genes, the AT content of all PCGs is 81.1%. It has a negative AT-skew and a positive GC-skew. The AT content of N-strand genes (75.7%) is higher than that of the J-strand genes (71.7%). The AT content of PCG third codon positions is much higher than that of the first and second codon positions and the AT content of PCG third codon positions of N-strand genes (87.1%) is the highest. PCGs of J-strand genes have negative AT-skew and GC-skew. PCGs of N-strand genes have negative AT-skew and positive GC-skew. For RNA genes, The AT content of tRNA was 76.9% and the AT-skew and GC-skew are positive. The AT content of the 16S rRNA (80.7%) is slightly higher than that of the 12S rRNA (77%). They both have negative AT-skew and a positive GC-skew.
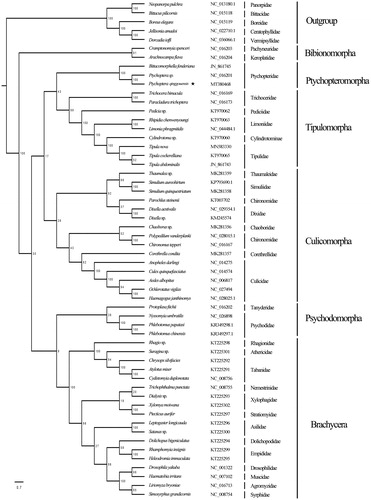
The phylogenetic tree in this study () shows a strong support for the monophyly of the Bibiomorpha (100%), Tipulomorpha (99%) and the Brachycera (100%). The phylogenetic relationship in Tipulomorpha is Trichoceridae + (Pediciidae + (Limoniidae + (Cylindrotomidae + Tipulidae))). The monophyly of the family Ptychopteridae is strongly supported, whereas its phylogenetic position in the lower diptera is undefined.
Acknowledgements
We express our sincere thanks to Ding Yang (Beijing) and Fan Song (Beijing) for great help during the study. We are also very grateful to Xin Li (Beijing) for collecting the specimen.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/. GeneBank accession numbers are listed as follows: Aedes albopitus (NC_006817), Anopheles darlingi (NC_014275), Arachnocampa flava (NC_016204), Atylotus miser (KT225291), Bittacomorphella fenderiana (JN_861745), Bittacus pilicornis (NC_015118), Boreus elegans (NC_015119), Chaoborus sp. (MK281356), Chironomus tepperi (NC_016167), Chrysops silvifacies (KT225292), Corethrella condita (MK281357), Cramptonomyia spenceri (NC_016203), Culex quinquefasciatus (NC_014574), Cydistomyia duplonotata (NC_008756), Cylindrotoma sp. (KT970060), Dialysis sp. (KT225293), Dixella aestivalis (NC_029354.1), Dixella sp. (KM245574), Dolichopus bigeniculatus (KT225294), Dorcadia ioffi (NC_036066.1), Drosophila yakuba (NC_001322), Haemagogus janthinomys (NC_028025.1), Haematobia irritans (NC_007102), Heleodromia immaculata (KT225295), Jellisonia amadoi (NC_022710.1), Leptogaster longicauda (KT225296), Limonia phragmitidis (NC_044484.1), Liriomyza bryoniae (NC_016713), Neopanorpa pulchra (NC_013180.1), Nyssomyia umbratilis (NC_026898), Ochlerotatus vigilax (NC_027494), Paracladura trichoptera (NC_016173), Parochlus steinenii (KT003702), Pedicia sp. (KT970062), Phlebotomus chinensis (KR349297.1), Phlebotomus papatasi (KR349298.1), Polypedilum vanderplanki (NC_028015.1), Protoplasa fitchii (NC_016202), Ptecticus aurifer (KT225297), Ptychoptera qinggouensis (MT380468), Ptychoptera sp. (NC_016201), Rhagio sp. (KT225298), Rhamphomyia insignis (KT225299), Rhipidia chenwenyoungi (KT970063), Satanas sp. (KT225300), Simosyrphus grandicornis (NC_008754), Simulium aureohirtum (KP793690.1), Simulium quinquestriatum (MK281358), Suragina sp. (KT225301), Thaumalea sp. (MK281359), Tipula abdominalis (JN_861743), Tipula cockerelliana (KT970065), Tipula nova (MN583330), Trichocera bimacula (NC_016169), Trichophthalma punctata (NC_008755) and Xylomya moiwana (KT225302).
Additional information
Funding
References
- Alexander CP. 1916. New or little-known crane-flies from the United States and Canada: Tipulidae, Ptychopteridae, Diptera. Part 3. Proc Acad Nat Sci Philadelphia. 68(3):486–549.
- Beckenbach AT. 2012. Mitochondrial Genome Sequences of Nematocera (Lower Diptera): Evidence of Rearrangement following a Complete Genome Duplication in a Winter Crane Fly. Genome Biol Evol. 4(2):89–101.
- Cameron SL. 2014. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst Entomol. 39(3):400–411.
- Caravas J, Friedrich M. 2013. Shaking the Diptera tree of life: performance analysis of nuclear and mitochondrial sequence data partitions. Syst Entomol. 38(1):93–103.
- Gillett C, Alex CP, Timmermans M, Jordal BH, Emerson BC, Vogler AP. 2014. Bulk de novo mitogenome assembly from pooled total DNA elucidates the phylogeny of weevils (Coleoptera: Curculionoidea). Mol Biol Evol. 31(8):2223–2237.
- Kang Z, Yao G, Yang D. 2013a. New species and record of Ptychoptera Meigen, 1803 (Diptera: Ptychopteridae) from China. Zootaxa. 3682(4):455–472.
- Kang ZH, Yao G, Yang D. 2013b. Five new species of Ptychoptera Meigen with a key to species from China (Diptera: Ptychopteridae). Zootaxa. 3682(4):541–555.
- Li H, Shao R, Song N, Song F, Jiang P, Li Z, Cai W. 2015. Higher-level phylogeny of paraneopteran insects inferred from mitochondrial genome sequences. Sci Rep. 5:8527.
- Li H, Shao R, Song F, Zhou X, Yang Q, Li Z, Cai W. 2013. Mitochondrial genomes of two Barklice, Psococerastis albimaculata and Longivalvus hyalospilus (Psocoptera: Psocomorpha): contrasting rates in mitochondrial gene rearrangement between major lineages of Psocodea. PLOS One. 8(4):e61685.
- Meigen JW. 1803. Versuch einer neuen Gattungs-Eintheilung der europaischen zweiflugligen Insekten. Magazin Für Insektenkunde (Illiger). 2:259–281.
- Paramonov NM. 2013. Ptychopteridae, a family of flies (Diptera) new to the philippine islands with the description of a new species. Zootaxa. 3682 (4):584–588.
- Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. 2006. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 37(1):545–579.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Timmermans MJ, Vogler AP. 2012. Phylogenetically informative rearrangements in mitochondrial genomes of Coleoptera, and monophyly of aquatic elateriform beetles (Dryopoidea). Mol Phylogenet Evol. 63(2):299–304.
- Török E, Kolcsár LP, Dénes AL, Keresztes L. 2015. Morphologies tells more than molecules in the case of the European widespread Ptychoptera albimana (Fabricius, 1787) (Diptera, Ptychopteridae). North-western. J Zool. 11 (2):304–315.
- Wang Y, Liu X, Garzón-Orduña IJ, Winterton SL, Yan Y, Aspöck U, Aspöck H, Yang D. 2017. Mitochondrial phylogenomics illuminates the evolutionary history of Neuropterida. Cladistics. 33(6):617–636.
- Wang Y, Liu X, Winterton SL, Yang D. 2012. The first mitochondrial genome for the fishfly subfamily Chauliodinae and implications for the higher phylogeny of megaloptera. PLOS One. 7(10):e47302.
- Westwood JO. 1835. Insectorum novorum exoticorum (ex ordine Dipterorum) descriptiones. London Edinburgh Philos Mag J Sci. 6(34):280–281.