Abstract
Viola Prionantha belongs to the family Violaceae. It has been widely used for a traditional Chinese herb with antibacterial activity and is grown as an early spring ornamental species in China. In this study, we determined the complete chloroplast genome sequence of V. prionantha which forms a circular structure. The whole chloroplast genome was 156,501 bp in length, consisting of a pair of inverted repeats (IR) of 26,404 bp, a large single-copy (LSC) region of 85,689 bp, and a small single-copy (SSC) region of 18,004 bp. We annotated 131 genes, including 84 coding sequences, 8 rRNA sequences, 37 tRNA sequences and 2 pesudogenes. Among the annotated genes, 17 genes contained one or two introns. Furthermore, a phylogenetic analysis revealed that V. prionantha and V. seoulensis clustered together as sisters to other Violaceae species.
Viola Prionantha is a perennial herb which belongs to the genus Viola in the family Violaceae, distributed in China, North Korea and Siberia (Duan et al. Citation2004; Wang Citation2009; Kyeong et al. Citation2020; Cheon et al. Citation2019). Viola Prionantha is used as an ornamental plant and has medicinal value with antibacterial activity (Liu and Sun Citation2006; Zhou Citation2008). Viola L. is known as one of the more difficult groups to classify. The phylogenetic relationships are still unclear among the genus (Yockteng et al. Citation2003; Liang and Xing Citation2010). In recent years, some studies have been conducted on the resources, medicinal properties, physiological characteristics, tissue culture in vitro, plant regeneration and heavy metal enrichments of the species (Zhang et al. Citation2012, Citation2013; Li et al. Citation2015; Zhao et al. Citation2016), but there have been no reports on its whole chloroplast genome. cpDNA (chloroplast DNA) is present in the mesophyll cells of green plants. In this study, we sequenced, assembled, and annotated the chloroplast genome for further studies on the phylogenomics of V. Prionantha.
The sampled V. prionantha fresh leaves were collected from Luoyang (34○64039.100N, 112○38080.700E), Henan province, in China. Voucher specimen (no. haust69l09) was deposited in the Henan University of Science and Technology. The total DNA was extracted according to a modified CTAB method (Doyle and Doyle Citation1987). The extracted Genomic DNA was used for sequencing with the Illumina NovaSeq platform. The reference genome of V. seoulensis (GenBank accession number: KP749924) and the programs such as SPAdes (Bankevish et al. Citation2012) and CpGAVAS (Liu et al. Citation2012) were used for sequence assembly and annotation. The cpDNA physical map was drawn using the OGDRAW tool (Greiner et al. Citation2019). Moreover, the complete chloroplast genome sequence was deposited in the GenBank database and a phylogenetic tree was constructed.
The plastid genome of V. prionantha (GenBank accession no. MT610374) forms a circular structure comprising 156,501 bp in length with 36.29% GC content, consisting of a pair of inverted repeats (IR) of 26,404 bp, a large single-copy (LSC) region of 85,689 bp, and a small single-copy (SSC) region of 18,004 bp. We annotated 131 genes, which consisted of 84 coding sequences, 2 pesudogenes, 8 rRNAs, and 37 tRNAs. Among the annotated genes, 17 genes contain one or two introns.
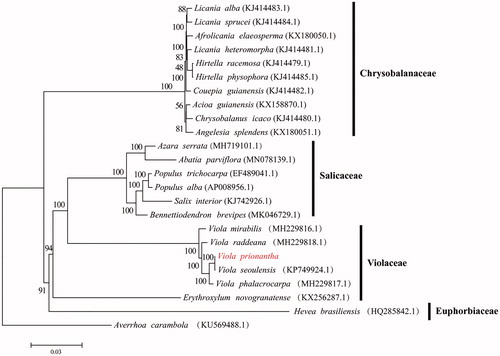
We conducted a phylogenetic analysis. A phylogenetic tree was constructed based on the following chloroplast genomes (accession number in parentheses) (). The genome sequences were aligned with MAFFTv7.427 (Katoh and Standley Citation2013) and then the maximum-likelihood (ML) tree was conducted using RAxMLv.8.2.10 with 1000 boot- strap replicates and the GTRGAMMA model (Stamatakis Citation2006). The phylogenetic analysis indicated V. prionantha was closely related to V. seoulensis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available NCBI (the National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov/Genbank, accession number MT610374.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, et al. 2012. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J Comput Biol.
- Cheon KS, Kim KA, Kwak M, Lee B, Yoo K. 2019. The complete chloroplast genome sequences of four Viola species (Violaceae) and comparative analyses with its congeneric species. PLOS One. 14(3):e0214162
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Duan CY, Hou XG, Zhang ZP, Li LF. 2004. Study on garden utilizes of Viola in China. Chin Agri Sci Bull. 20(5):185–186.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kyeong SC, Dong KK, Kyung AK, Ki OY. 2020. The complete chloroplast genome sequence of Viola japonica (Violaceae). Mitochondrial DNA Part B. 5(2):1297–1298.
- Li JQ, Lin LH, Zhang F, Wan XQ, Liu M, Zhao JL. 2015. Tissue culture in vitro and plant regeneration of Viola prionantha. Acta Prataculturae Sin. 24(11):163–173.
- Liang GX, Xing FW. 2010. Infrageneric phylogeny of the genus Viola (Violaceae) based on trnL-trnF, psbA-trnH, rpl16, ITS sequences, cytological and morphological data. Acta Botanica Yunnanica. 32:477–488.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Liu HC, Sun ZY. 2006. Study on the introduction and application of three kinds of wild genus Viola. Chin Landscape Architec. 1(13):1–4.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Wang LN. 2009. The life cycle and ecological adaptation of V. Prionantha Bunge. Hohhot: Inner Mongolia University.
- Yockteng R, Ballard HE, Mansion G, Dajoz I, Nadot S. 2003. Relationships among pansies (Viola section Melanium) investigated using ITS and ISSR markers. Plant Systemat Evol. 241(3–4):153–170.
- Zhang C, Xu X, Li XK, Yan H, Dong L. 2012. Investigation and analysis of native herbaceous ground cover plants in Beijing Olympic Forest Park. Pratacult Sci. 29(8):1193–1198.
- Zhang F, Wan XQ, Liu M, Zhong Y, Liu GL, Zhang S, Luo B, Huang JL, Wu FY. 2013. Method of tissue culture and rapid propagation of Viola prionantha. Chinese Patent CN103004611A, 2013-04-03.
- Zhao JL, Zhang F, Wan XQ, Xiao Z. 2016. Cadmium tolerance and enrichment characteristics of Viola prionantha. Pratacult Sci. 33(1):54–60.
- Zhou C. 2008. The textual research and the studies on the chemical constituent of Viola yedoensis. Beijing: Beijing University of Chinese Medicine.