Abstract
Lepturacanthus savala is a commercially important marine fish belonging to the family Trichiuridae, with a wide distribution in the Indian-Western Pacific coastal waters. In this study, the complete mitochondrial genome of L. savala has been determined by primer walking methods. The mitogenome is 17,146 bp in length containing 13 protein-coding genes, 2 ribosomal RNA genes, 21 transfer RNA genes, a control region, and an origin of light-chain replication region. The base composition is 28.9% for A, 29.0% for C, 15.7% for G, and 26.4% for T. The maximum-likelihood tree based on whole mitogenome showed that it is the closest related species of Trichiurus.
The genus Lepturacanthus Fowler, 1905 belongs to the family Trichiuridae and comprises three valid species: Lepturacanthus savala (Cuvier, 1829), Lepturacanthus pantului (Gupta, 1966), and Lepturacanthus roelandti (Bleeker, 1860), features very elongated and strongly compressed body with a ribbon-like caudal fin, gradually tapering to a point (Nakamura and Parin Citation1993; Chakraborty et al. Citation2006). In this study, we present the complete mitochondrial genome of L. savala, which has been deposited in GenBank with accession no. MT269921.
Samples were collected from Qingdao, Yellow Sea (120°46.064′, 36°22.452′) in 10 November 2017. Specimens were preserved in 95% ethanol and deposited at the Fish Specimen Room of Fisheries Ecology Laboratory (specimen accession no. OUC_FEL201910051), Fisheries College, Ocean University of China, Qingdao City, Shandong Province, China. Total genomic DNA was isolated from the muscle tissue by proteinase K digestion followed by the standard phenol–chloroform method (Sambrook and Russell Citation2001). PCR primer sets were designed using Primer Premier 6.0 (PREMIER Biosoft International, Palo Alto, CA) based on the complete mitochondrial genome sequences of Trichiurus nanhaiensis (GenBank accession no. NC018791) and Trichiurus japonicus (GenBank accession no. MK292708). The normal PCR was performed following the standard procedure (Liu et al. Citation2006). The tRNA genes were predicted using online software tRNAScan-SE (Lowe and Chan Citation2016).
The complete mitogenome of L. savala is 17,146 bp in length, containing 13 protein-coding genes, 21 tRNA genes, 2 rRNA genes, a control region (D-Loop), and an origin of light strand replication (OL). Different with typical 22 tRNA genes in vertebrate mitogenome, the mitogenome contained 21 tRNA genes except for tRNAPro. The tRNAPro gene is absent also in T. japonicus and T. nanhaiensis (Liu and Cui Citation2009; Liu et al. Citation2013; Xu et al. Citation2019; Zheng et al. Citation2019). Most of the genes are transcribed on the heavy strand (H-strand), except for seven tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser-UGA, Glu). The base composition is 28.9% for A, 29.0% for C, 15.7% for G, and 26.4% for T, with a slight AT bias of 55.3%. The small non-coding region, a light-chain replication region (OL) was observed between the tRNAAsn and tRNACys in WANCY region and it could form a stable stem-loop secondary structure. The largest non-coding region (control region) located between the tRNAThr and tRNAPhe genes is determined to be 1485 bp in length. Within the control region, termination associated sequence (TAS), central conserved domain (CSB-D, E, F), conserved sequence block (CSB-1, 2, 3), as well as two different types of tandem repeat sequences were detected respectively.
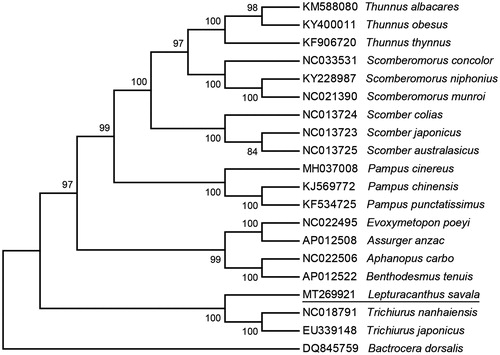
Maximum-likelihood phylogeny of L. savala and other 17 Scombriformes fishes based on complete mitochondrial genomes were reconstructed, using Bactrocera dorsalis (DQ845759) as an outgroup in the MEGA 7 software (Kumar et al. Citation2016). This phylogenetic tree shows that L. savala is the closest related species of Trichiurus with high bootstrap value supported ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, accession number [MT269921].
Additional information
Funding
References
- Chakraborty A, Oijen M, Lim KKP, Iwatsuki Y. 2006. Lepturacanthus roelandti (Bleeker, 1860), a valid species of hairtail (Perciformes: Trichiuridae). Ichthyol Res. 53(1):41–46.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu J, Gao T, Wu S, Zhang Y. 2006. Pleistocene isolation in the northwestern Pacific marginal seas and limited dispersal in a marine fish, Chelon haematocheilus (Temminck & Schlegel, 1845). Mol Ecol. 16(2):275–288.
- Liu X, Guo Y, Wang Z, Liu C. 2013. The complete mitochondrial genome sequence of Trichiurus nanhaiensis (Perciformes: Trichiuridae). Mitochondrial DNA. 24(5):516–517.
- Liu Y, Cui Z. 2009. The complete mitochondrial genome sequence of the cutlassfish Trichiurus japonicus (Perciformes: Trichiuridae): genome characterization and phylogenetic considerations. Mar Genomics. 2(2):133–142.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: search and contextual analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Nakamura I, Parin NV. 1993. FAO species catalogue, snake mackerels and cutlassfish of the world (Families Gempylidae and Trichiuridae). FAO Fish Catalogue. 125:99–101.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. 3rd ed. New York: Cold Spring Harbor Laboratory Press.
- Xu L, Wang X, Du F. 2019. The complete mitochondrial genome of cutlassfish (Trichiurus japonicus) from South China Sea. Mitochondrial DNA Part B. 4(1):783–784.
- Zheng S, Liu J, Jia P. 2019. Complete mitogenome of the cutlassfish Trichiurus haumela (Scombriformes: Trichiuridae) from Ningde, Fujian Province, Southeast China. Mitochondrial DNA Part B. 4(1):87–88.