Abstract
Michelia champaca var. champaca is an ornamentally important tree in Magnoliaceae. The paper reported the complete chloroplast genome (cpDNA) of M. champaca var. champaca and its basic annotated information. The size of cpDNA is 160,008 bp, with a typical quadripartite structure of a large single-copy (LSC) region of 88,037 bp and a small single-copy (SSC) region of 18,809 bp separated by a pair identical inverted repeat regions (IRs) of 26,581 bp each. The genome contained 131 genes (113 unique), including 86 protein-coding genes (80 unique), 37 tRNA genes (29 unique), and eight rRNA genes (four unique). Phylogenetic analysis showed that M. champaca var. champaca is affinal to M. baillonii and they form a nomophyletic group with other eight Michelia species. This Michelia clade is sister to the Aromadendron cathcartii clade with high support. All genera mentioned in this analysis are nomophyletic under the system of Magnoliaceae by Sima and Lu.
Michelia champaca Linnaeus is the type species of the genus Michelia Linnaeus in the family Magnoliaceae (Sima and Lu Citation2012). It has got two varieties, var. champaca Linnaeus and var. pubinervia (Blume) Miquel. The latter variety occurs in China (S Xizang, S and SW Yunnan), Cambodia, India, Indonesia, Laos, Malaysia, Myanmar, Nepal, Thailand, and Vietnam (Xia et al. Citation2008; Nooteboom and Chalermglin Citation2009). Michelia champaca var. champaca Linnaeus is distributed in China (SE Xizang, S and SW Yunnan), Bangladesh, India, Malaysia, Myanmar, Nepal, Pakistan, and Sri Lanka and widely cultivated as a good ornamental species in Southern China and Southeastern Asia, which is probably originally from India (Xia et al. Citation2008; Kundu Citation2009; Nooteboom and Chalermglin Citation2009; Jiang et al. Citation2013). It is grown as a plant for perfume and used medicinally (Khan et al. Citation2002; Jiang et al. Citation2012; Ma et al. Citation2012; Sinha and Varma Citation2017). Its flowers are used as nice perfume trinkets by women in many places in China (Jiang et al. Citation2013). However, there has been no report on chloroplast genome information of M. champaca var. champaca Linnaeus until now.
The complete sequence of chloroplast genome of M. champaca var. champaca Linnaeus was reported in this study. Genomic DNA was extracted using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA, USA) from the fresh leaves of M. champaca var. champaca Linnaeus collected from Kunming Arboretum, Yunnan Academy of Forestry and Grassland Science, Yunnan Province of China (25°9'8″ N, 102°44'46″ E). The sheets of vouchered specimen (Y. K. Sima and S. Y. Chen 99278) are deposited at the herbaria, YAF and YCP.
Total genome DNA of M. champaca var. champaca Linnaeus was sequenced by Illumina HiSeq Sequencing System (Illumina, San Diego, CA) and shotgun library was constructed. About 2.0 G pair-end (150 bp) raw sequence data were obtained and the low-quality sequences were filtered through CLC Genomics Workbench v8.0 (CLC Bio, Aarhus, Denmark) to get high-quality clean reads. NOVO Plasty software (Dierckxsens et al. Citation2017) was used to align and assemble cp genome with Pachylarnax sinica (Y. W. Law) N. H. Xia and C. Y. Wu (JX280400) served as the reference. The genome was automatically annotated using CpGAVAS (Liu et al. Citation2012) and then adjusted and confirmed with Geneious 9.1 (Kearse et al. Citation2012). The annotated genomic sequence was submitted to GenBank under Accession Number of MT269873 (https://www.ncbi.nlm.nih.gov/nuccore/MT269873.1). To better determine the phylogenetic position of M. champaca var. champaca Linnaeus, the complete chloroplast genome sequences of other thirty-five species of the subfamily Magnolioideae from NCBI were aligned using MAFFT v. 7 (Sima and Lu Citation2012; Katoh and Standley Citation2013; Sima et al. Citation2014). Based on the system of Magnoliaceae by Sima and Lu (Citation2012), two species of the subfamily Liriodendroideae, Liriodendron chinense (Hemsley) Sargent (KU170538) and Liriodendron tulipifera Linnaeus (DQB99947) were served as the outgroup. The maximum likelihood (ML) tree was reconstructed with RAxML (implemented in Geneious ver.10.1 http://www.geneious.com, Kearse et al. Citation2012) and bootstrap probability values were calculated from 1000 replicates.
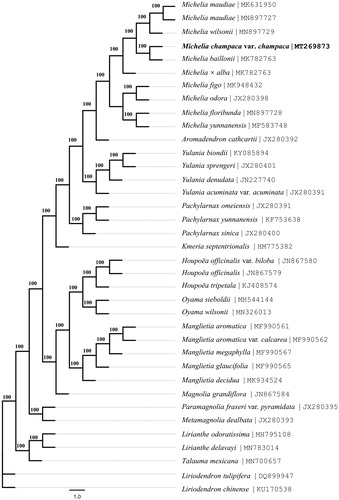
The complete cp genome of M. champaca var. champaca Linnaeus is 160,008 bp in length, with a large single-copy (LSC) region of 88,037 bp and a small single-copy (SSC) region of 18,809 bp separated by a pair of identical inverted repeat regions (IRs) of 26,581 bp each. The cp genome contained 131 genes (113 unique), including 86 protein-coding genes (80 unique), 37 tRNA genes (29 unique), and eight rRNA genes (four unique). The result of phylogenetic analysis showed that M. champaca var. champaca Linnaeus is affinal to Michelia baillonii (Pierre) Finet and Gagnepain (MK782763) and they form a nomophyletic group with other eight species of the genus Michelia Linnaeus (). This clade of the genus Michelia Linnaeus is sister to the clade of Aromadendron cathcartii (J. D. Hooker and Thomson) Sima and S. G. Lu with high support. All genera mentioned in this analysis are nomophyletic under the system of Magnoliaceae by Sima and Lu (Citation2012). The study would provide effective new molecular data for evolutionary and phylogenetic analysis of Magnoliaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The sheets of vouchered specimen, Y. K. Sima and S. Y. Chen 99278, have been stored at the herbaria, YAF and YCP.
The data of complete chloroplast genome sequence of Michelia champaca var. champaca Linnaeus have been deposited into NCBI databases and the accession number is MT269873 (https://www.ncbi.nlm.nih.gov/nuccore/MT269873.1).
The data above should only be shared if it is ethically correct to do so, where this does not violate the protection of human subjects, or other valid ethical, privacy, or security concerns.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. Novoplasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Jiang DY, Li YH, Xia B, He F, Pan HT. 2012. Components and antibacterial functions of volatile organic compounds from leaves and flowers of Michelia champaca. J Northeast Forest Univ. 40(5):71–74.
- Jiang DY, Shen X, Li YH, Pan HT. 2013. Physiological and biochemical characteristics of Michelia champaca L. during florescence and flower senescence. Chinese Agri Sci Bull. 29(16):125–128.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Khan MR, Kihara M, Omoloso AD. 2002. Antimicrobial activity of Michelia champaca. Fitoterapia. 73(7-8):744–748.
- Kundu SR. 2009. A synopsis on distribution and endemism of Magnoliaceae s.l. in Indian Subcontinent. Thaiszia. 19(1):47–60.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Ma HF, Sima YK, Hao JB, Chen SY, Han MY, Li D, Xu L, Zhou B, Chai Y. 2012. The chemical constituents in the volatile oils from Michelia champaca Linn. Fine Chemic. 29 (1):41–44. 56.
- Nooteboom HP, Chalermglin P. 2009. The Magnoliaceae of Thailand. Thai Bull (Bot). 37:111–138.
- Sima YK, Lu SG. 2012. A new system for the family Magnoliaceae. In: Xia NH, Zeng QW, Xu FX, Wu QG, editors. Proceedings of the second international symposium on the family Magnoliaceae. Wuhan: Huazhong University Science & Technology Press. p. 55–71.
- Sima YK, Lu SG, Hao JB, Xu T, Han MY, Xu L, Li D, Ma HF, Chen SY. 2014. Notes on Manglietia aromatica var. calcarea (Magnoliaceae), a variety endemic to South-Western China. J West China Forest Sci. 43(1):96–98.
- Sinha R, Varma R. 2017. Antioxidant activity in leaf extracts of Michelia champaca L. J Adv Pharm Edu Res. 7(2):86–88.
- Xia NH, Liu YH, (Law YW), Nooteboom HP. 2008. Magnoliaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China, Vol. 7. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press. p. 48–91.