Abstract
Senna spectabilis (DC.) H.S. Irwin & Barneby is a popular ornamental tree as well as a traditional medical plant in Cameroon. In this study, we sequenced and annotated the complete chloroplast genome of S. spectabilis and reconstructed the phylogenetic relationship of the tribe Cassieae. The length of the chloroplast genome was determined to be 162,754 bp, containing a pair of inverted repeats of 25,413 bp which separated by a small single-copy (SSC) region of 20,161 bp and a large single-copy (LSC) region of 91,767 bp. The cp genome encodes 128 genes, including 83 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The percentage of total GC content of this genome was 35.7%. The phylogenetic analysis indicates that S. spectabilis with the sampled Senna species formed a well-supported monophyletic clade.
Senna spectabilis (DC.) H.S. Irwin & Barneby is a popular ornamental tree with bright yellow and fragrant flowers, which is native to tropical America and widely cultivated in south Yunnan and Guangdong Province of China (Chen et al. Citation2010). Moreover, S. spectabilis was also used as medical plant in Cameroon to treat anxiety, constipation, epilepsy, and insomnia (Nkantchoua et al. Citation2018). The leaves, flowers, and fruits of S. spectabilis are important source of cytotoxic alkaloids (de Albuquerque Melo et al. Citation2014). However, the phylogenetic relationship of the genus Senna and the phylogenetic position of S. spectabilis are still unresolved. In the present study, we report the first complete chloroplast genome of S. spectabilis and perform the phylogenetic analysis of the tribe Cassieae employ chloroplast phylogenomics.
Total genomic DNA was isolated from ca. 15 mg silica gel dried leaves following the modified CTAB method (Doyle and Doyle Citation1987). Leaf sample of S. spectabilis was collected from a cultivated tree in Experimental Station of Research Institute of Tropical Forestry, Chinese Academy of Forestry, Ledong County (18°41.651′N, 108°47.135′E), Hainan Province, China. The voucher specimens (accession number: S200009) were deposited at the herbarium of South China Botanical Garden (IBSC). Paired-end reads (PE = 150 bp) were produced using a BGISEQ-500 platform at Beijing Genomics Institute (Shenzhen, China). Before assembly, the raw reads were filtered by the program Trimmomatic v.0.33 (Bolger et al. Citation2014) with default parameters. Then the trimmed reads were assembled into complete chloroplast genome by using NOVOPlasty 2.6.3 (Dierckxsens et al. Citation2017) with kmer length set to 59 base pairs. Annotation of the cp genome was performed on Geneious version 11.0.3 (Kearse et al. Citation2012), while the tRNA genes were annotated on ARAGORN (Laslett and Canback Citation2004). The annotated plastome of S. obtusifolia (GenBank accession number: MK817504) was used as a reference for assembly and annotation.
The complete cp genome of S. spectabilis (GenBank accession number: MT635191) was 162,754 bp in length, containing a small single-copy region (SSC) of 20,161 bp, a large single-copy region (LSC) of 91,767 bp and a pair of inverted repeats (IRs) of 25,413 bp. The GC contents of this genome is 35.7%, while the GC contents of LSC, SSC, and IRs are 33.1%, 29.3%, and 42.9%, respectively. A total of 128 unique genes were identified from this genome, including 83 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Among these genes, 15 genes (trnK-UUU, rps16, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, ndhB, trnI-GAU, trnA-UGC, and ndhA) have one intron and three genes (clpP, rps12, and ycf3) have two introns.
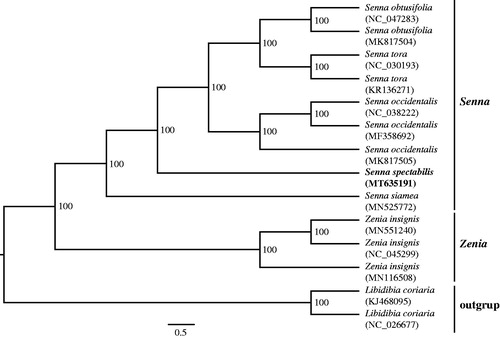
To identify the phylogenetic position of S. spectabilis, we reconstructed the phylogeny of the tribe Cassieae. The matrix includes all of the reported plastomes of this tribe and an outgroup from the genus Libidibia. The plastomes of the 14 accessions were aligned using MAFFT (Katoh and Standley Citation2013). Then the phylogenetic analysis was performed by using RAxML (Stamatakis Citation2006) with 1000 bootstrap replicates. The results support the monophyly of the genus Senna (). Senna spectabilis is sister to the clade which including S. obtusifolia, S. tora and S. occidentalis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT635191.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Chen DZ, Zhang DX, Larsen K. 2010. Cassieae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China, Vol. 10. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press. p. 27–34.
- de Albuquerque Melo GM, Silva MCR, Guimarães TP, Pinheiro KM, da Matta CBB, de Queiroz AC, Pivatto M, da Silva Bolzani V, Alexandre-Moreira MS, Viegas C Jr. 2014. Leishmanicidal activity of the crude extract, fractions and major piperidine alkaloids from the flowers of Senna spectabilis. Phytomedicine. 21(3):277–281.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16.
- Nkantchoua GCN, Njapdounke JSK, Fifen JJ, Taiwe GS, Ojong LJ, Kandeda AK, Bum EN. 2018. Anticonvulsant effects of Senna spectabilis on seizures induced by chemicals and maximal electroshock. J Ethnopharmacol. 212:18–28.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.