Abstract
Brechemia lineata is a well-known medicinal plant in the Rhamnaceae family and widely used in traditional Chinese medicine. Here, we sequenced the complete chloroplast genome using Illumina pair-end sequencing data. The chloroplast genome was 154,962 bp in length, consisting of a large single-copy (LSC) region of 82,928 bp, a small single-copy (SSC) region of 17,376 bp, and a pair of inverted repeat (IR) regions of 27,329 bp. The chloroplast genome consists of 112 unique genes, including 78 protein-coding genes, 30 transfer RNA, and 4 ribosomal RNA genes. The overall GC content of the chloroplast genome was 37.0%. The phylogenetic analysis suggests close relationship of B. lineata with other Berchemia species. These genomic resources will be valuable resource for systematic and phylogenetic studies of Berchemia genus.
Berchemia is a genus of Rhamnaceae family, comprises of about 32 species and mainly distributed in temperate and tropical areas of East to Southeast Asia (Chen and Carsten Citation2007). Berchemia lineata (L.) DC. is a traditional Chinese medicinal species distributed in China (Fujian, Guangdong, Guangxi and Hainan), Japan and Vietnam. Berchemia lineata roots and leaves are used medicinally for relieving coughs and reducing sputum and for treating injuries, trauma, and snakebites (Chen and Carsten Citation2007). Berchemia species are often misidentified due to extremely similar vegetative morphological characteristics (Guo et al. Citation2016). Chloroplast genome has been broadly used for reconstructing phylogenetic relationships and development of molecular maker for the identification of plant species (Jansen et al. Citation2007; Huang et al. Citation2020). In this study, we sequenced the chloroplast genome of B. lineata to provide valuable genomic resources to facilitate systematic and phylogenetic studies of this important medicinal plant.
Berchemia lineata fresh leaves were collected from Zengcheng district in Guangzhou city (Guangdong, China: N23°13′49.0″, E113°45′45.2″) and voucher specimen (Zhou2019-18) was deposited in the Herbarium of Sun Yat-sen University (SYS). Genomic DNA was isolated using Tiangen plant genomic DNA kits (Tiangen Biotech, Beijing) and sequenced on the Illumina Hi-Seq 2500 platform. GetOrganelle pipeline were used for de novo chloroplast genome assembly (Jin et al. Citation2019). The chloroplast genome was annotated using Geseq and then manually verified and visualized in Geneious Prime v.2019.1.3 (Kearse et al. Citation2012; Tillich et al. Citation2017). Finally, circular chloroplast genome map was drawn using OGDRAW (Lohse et al. Citation2013). The chloroplast genome was deposited in the GenBank with the accession number MT621210.
Berchemia lineata chloroplast genome had quadripartite structure with 154,962 bp in length, consisted of a large single-copy (LSC) region of 82,928 bp, a small single-copy (SSC) region of 17,376 bp, and a pair of inverted repeat (IR) regions of 27,329 bp. The chloroplast genome contained 112 unique genes, including 78 protein-coding genes, 30 transfer RNA genes, and 4 ribosomal RNA genes. The overall GC content of the chloroplast genome was 37.0% and the corresponding values of LSC, SSC, and IR regions were 34.7%, 31.2%, and 42.4%, respectively.
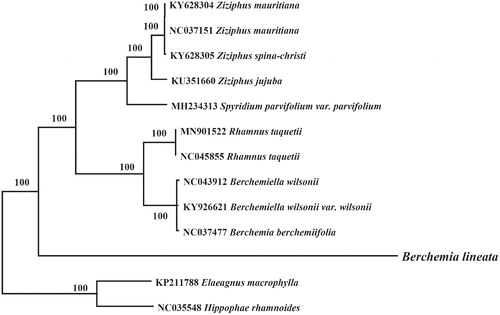
Phylogenetic relationship of B. lineata within the Rhamnaceae family was inferred using the previously published eight chloroplast genome from the Rhamnaceae family and Hippophae rhamnoides and Elaeagnus macrophylla (Elaeagnaceae) as out group. The chloroplast genome of these species were aligned using MAFFT v7.3 (Katoh and Standley Citation2013) and MEGA7 were used to construct neighbor-joining phylogenetic tree (Kumar et al. Citation2016). Phylogenetic analysis revealed that B. lineata clustered with Berchemia species, however making a separate clade () might be due to geographical isolation from B. berchemiifolia (only distributed in the Korean peninsula and Japan) (Cheon et al. Citation2018). This chloroplast genome of B. lineata will provide a fundamental resource for studying the systematic position and conservation of this important medicinal species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank at https://www.ncbi.nlm.nih.gov/, reference number MT621210.
Additional information
Funding
References
- Chen Y, Carsten S. 2007. Rhamnaceae. Flora of China. Beijing: Science Press.
- Cheon KS, Kim KA, Yoo KO. 2018. The complete chloroplast genome sequence of Berchemia berchemiifolia (Rhamnaceae). Mitochondrial DNA Part B. 3(1):133–134.
- Guo LC, Zhao MM, Sun W, Teng HL, Huang BS, Zhao XP. 2016. Differentiation of the Chinese minority medicinal plant genus Berchemia spp. by evaluating three candidate barcodes. Springerplus. 5(1):658–658.
- Huang R, Liang Q, Wang Y, Yang TJ, Zhang Y. 2020. The complete chloroplast genome of Epimedium pubescens Maxim. (Berberidaceae), a traditional Chinese medicine herb. Mitochondrial DNA Part B. 5(3):2042–2044.
- Jansen RK, Cai ZQ, Raubeson LA, Daniell H, Leebens-Mack J, Muller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 104(49):19369–19374.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2019. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv 4:256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–W581.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes . Nucleic Acids Res. 45(W1):W6–W11.