Abstract
Mahachai bettas (Betta mahachaiensis) are distributed in areas of brackish water with Nipa Palms in Samut Sakhon, Thailand but urbanization is restricting their biodiversity. A complete mitochondrial genome (mitogenome) of B. mahachaiensis was determined to support conservation programs. Mitogenome sequences were 16,980 bp in length with slight AT bias (61.91%), containing 37 genes with identical order to most teleost mitogenomes. Phylogenetic analysis of B. mahachaiensis showed a closer relationship with B. splendens. Results will allow the creation of a reference annotated genome that can be utilized to sustain biodiversity and eco-management of the betta to improve conservation programs.
Keywords:
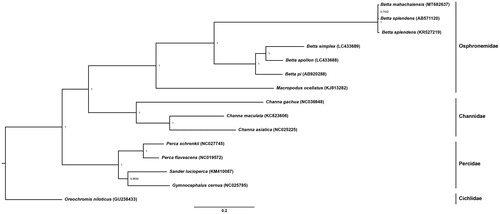
The location of the Southeast Asia (SEA) mainland and numerous islands, and its hot and humid climate provides a huge variety of bioresources. Fighting fish or betta (Betta spp.), which are native to SEA, have been proposed to study their fighting and ornamental attributes. Long-term selection as crossbreeding between different lines or species has honed their aggressive behavior and body features, resulting in large novel varieties (Witte and Schmidt Citation1992; Ramos and Gonçalves Citation2019). However, these widespread activities of artificial selection have resulted in inbreeding depression and outbreeding depression for hybrid betta between different species/lines. The large biodiversity of betta is currently being lost more rapidly than at any time in the past several million years, with the invasion of alien species or hybrids introduced into the wild leading to genetic admixture (Beer et al. Citation2019). This is a very serious problem in the context of conservation biology and genetics and needs to be resolved as a matter of urgency. Betta mahachaiensis is found in brackish waters with Nipa Palms in Samut Sakhon, Samut Songkhram, Samut Prakan, and Bangkok Provinces (Kowasupat et al. Citation2012). This species is now endangered as a result of infrastructural expansion through urbanization. Here, a complete mitochondrial genome (mitogenome) of B. mahachaiensis collected from Samut Sakhon (13.550333 N, 100.273968 E) was determined and stored in the Thailand Natural History Museum (No. THM21090). Whole genomic DNA was extracted in accordance with the standard salting-out protocol (Supikamolseni et al. Citation2015). Next-generation sequencing was performed using an Illumina HiSeq platform at Vishuo Biomedical (Thailand) Ltd. The quality of Illumina reads was evaluated with FASTQC and the raw reads were trimmed to discard adapters using trimmomatic (Bolger et al. Citation2014). The trimmed reads were subjected to alignments to isolate all mitogenome sequences by mapping whole genome Illumina reads against the complete mitogenome of B. splendens (AB571120), using bowtie2 – end-to-end and very-sensitive parameters (Langmead and Salzberg Citation2012). The mapped alignment was processed using SAMtools (Li et al. Citation2009), and aligned reads were extracted using BEDtools (Quinlan and Hall Citation2010). The aligned reads with the mitogenome were then de novo assembled using Velvet (Velvet_1.1.07; kmer = 91) (Zerbino and Birney Citation2008). A total of 63,643 individual reads gave an average coverage of around 300X. We also generated a consensus genome sequence of the read alignments from the variants against the reference using BCFtools (Li Citation2011) and VCFtools (Danecek et al. Citation2011) with annotations generated in the MITOS WebServer (Bernt et al. Citation2013). Complete mitogenome sequences consisted of 16,980 bp for B. mahachaiensis (GenBank Accession number: MT682637, BioProject: PRJNA642255), containing 37 genes and a control region (CR). Gene arrangement patterns were identical to those of teleosts (Miya et al. Citation2013). Overall AT content values for the mitogenome was 61.91%. Average nucleotide diversity among all Betta mitogenomes was determined at 20.43 ± 0.03%. Four conserved sequence blocks (CSB-D, CSB1, CSB2, and CSB3) in the CR of teleost mitogenomes were also present in B. mahachaiensis (Lee and Kocher Citation1995; Prakhongcheep et al. Citation2018; Ponjarat et al. Citation2019). Diverse numbers of tandem repeats were observed in B. pi (AB920288) and B. splendens (AB571120 and KR527219) (Song et al. Citation2016; Prakhongcheep et al. Citation2018; Ponjarat et al. Citation2019), suggesting that the CR had a large variation in different fighting fish species. A phylogenetic tree was constructed based on twelve concatenated protein-coding genes without ND6 of 15 teleosts, using Bayesian inference with MrBayes version 3.2.6 (Huelsenbeck and Ronquist Citation2001). The sister group comprising B. splendens and B. mahachaiensis formed a monophyletic clade, consistent with Sriwattanarothai et al. (Citation2010). The two species were also close to the other betta clade, confirming results from previous studies (Ruber et al. Citation2004) (). These complete mitogenomes will allow the creation of a reference annotated genome, and provide valuable information at the molecular level that can be utilized to sustain biodiversity and eco-management of the betta to improve conservation management.
Figure 1. Phylogenetic relationships among twelve concatenated mitochondrial protein-coding genes, without ND6 sequences of fifteen mitochondrial genomes, including, Oreochromis niloticus as the outgroup using Bayesian inference analysis. The complete mitochondrial genome sequence was downloaded from GenBank. Accession numbers are indicated in parentheses after the scientific names of each species. Support values at each node are Bayesian posterior probabilities, while branch lengths represent the number of nucleotide substitutions per site.
Acknowledgements
The authors would like to thank Sunchai Makchai (Thailand Natural History Museum) for advice on sample preparation. We are also indebted to Vishuo Biomedical (Thailand) Ltd. for excellent service collaboration. The Center for Agricultural Biotechnology (CAB) at Kasetsart University Kamphaeng Saen Campus provided support with server analysis services, and the Faculty of Science and the Faculty of Forestry at Kasetsart University supported the research facilities. The National Biobank of Thailand (NBT) under the National Science and Technology Development Agency (NSTDA), Thailand supported this study through the use of a high performance computer. We also thank the Plakad Association, Thailand for providing information concerning Betta spp. in Thailand.
Disclosure statement
The authors report no conflicts of interest and are entirely responsible for the contents of this article. Animal care and all experimental procedures were approved by the Animal Experiment Committee, Kasetsart University, Thailand (approval no. ACKU63-SCI-007) and conducted in accordance with the Regulations on Animal Experiments at Kasetsart University.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT682637.
Additional information
Funding
References
- Beer SD, Cornett S, Austerman P, Trometer B, Hoffman T, Bartron ML. 2019. Genetic diversity, admixture, and hatchery influence in Brook Trout (Salvelinus fontinalis) throughout western New York State. Ecol Evol. 9(13):7455–7479.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics. 30(15):2114–2120.
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics. 27(15):2156–2158.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Kowasupat C, Panijpan B, Ruenwongsa P, Sriwattanarothai N. 2012. Betta mahachaiensis, a new species of bubble-nesting fighting fish (Teleostei: Osphronemidae) from Samut Sakhon Province, Thailand. Zootaxa. 3522(1):49–60.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Lee WJ, Kocher TD. 1995. Complete sequence of a sea lamprey (Petromyzon marinus) mitochondrial genome: early establishment of the vertebrate genome organization. Genetics. 139(2):873–887.
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 27(21):2987–2993.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Miya M, Friedman M, Satoh TP, Takeshima H, Sado T, Iwasaki W, Yamanoue Y, Nakatani M, Nakatani K, Inoue JG, et al. 2013. Evolutionary origin of the Scombridae (Tunas and Mackerels): members of a paleogene adaptive radiation with 14 other pelagic fish families. PLoS One. 8(9):e73535.
- Ponjarat J, Areesirisuk P, Prakhongcheep O, Dokkaew S, Sillapaprayoon S, Muangmai N, Peyachoknagul S, Srikulnath K. 2019. Complete mitochondrial genome of two mouthbrooding fighting fishes, Betta apollon and B. simplex (Teleostei: Osphronemidae). Mitochondrial DNA B Resour. 4:672–674.
- Prakhongcheep O, Muangmai N, Peyachoknagul S, Srikulnath K. 2018. Complete mitochondrial genome of mouthbrooding fighting fish (Betta pi) compared with bubble nesting fighting fish (B. splendens). Mitochondrial DNA B Resour. 3(1):6–8.
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26(6):841–842.
- Ramos A, Gonçalves D. 2019. Artificial selection for male winners in the Siamese fighting fish Betta splendens correlates with high female aggression. Front Zool. 16:34.
- Ruber L, Britz R, Tan HH, Ng PKL, Zardoya R. 2004. Evolution of mouthbrooding and life-history correlates in the fighting fish genus Betta. Evolution. 58(4):799–813.
- Song YN, Xiao GB, Li JT. 2016. Complete mitochondrial genome of the Siamese fighting fish (Betta splendens). Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4580–4581.
- Sriwattanarothai N, Steinke D, Ruenwongsa P, Hanner R, Panijpan B. 2010. Molecular and morphological evidence supports the species status of the Mahachai fighter Betta sp. Mahachai and reveals new species of Betta from Thailand. J Fish Biol. 77(2):414–424.
- Supikamolseni A, Ngaoburanawit N, Sumontha M, Chanhome L, Suntrarachun S, Peyachoknagul S, Srikulnath K. 2015. Molecular barcoding of venomous snakes and species-specific multiplex PCR assay to identify snake groups for which antivenom is available in Thailand. Genet Mol Res. 14(4):13981–13997.
- Witte K, Schmidt J. 1992. Betta brownorum, a new species of anabantoids (Teleostei; Belontiidae) from northwestern Borneo, with a key to the genus. Ichthyol Explor Freshw. 2:305–330.
- Zerbino D, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
