Abstract
The complete mitochondrial genome of a horntail wasp, Tremex fuscicornis, was analyzed prior to a systematic study on South Korean Siricidae. This species genome has a total length of 16,359 bp (GenBank accession number: MT425058), consisting of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and an A + T rich control region. The nucleotide composition is 41.8% T, 38.2% A, 14.3% C, and 5.7% G. The result of the maximum-likelihood analysis indicates that T. fuscicornis is a distinctive species in Siricidae, and this mitogenomic sequence can be used as a reference for phylogenetic studies on the superfamily Siricoidea.
Hymenoptera is one of the largest insect orders, and contains over 150,000 species with various life strategies. Up to date, only about 270 hymenopteran species have been presented with complete or nearly complete mitochondrial genomes (Yan et al. Citation2019). The members of Siricidae are common but serious wood-boring insect pests and commonly known as horntails or wood wasps. Siricidae is mainly distributed in the Northern Hemisphere, containing 111 extant species in 11 genera worldwide, and 14 species in five genera are known from Korea (Aguiar et al. Citation2013; National Institute of Biological Resources Citation2019).
Females of some Tremex species inoculate their tree hosts with the symbiotic basidiomycete wood-decaying fungi at oviposition. The tunneling by hatched larvae and wood-rotting by fungi make this insect–fungus mutualism harmful to host trees (Pažoutová and Šrůtka Citation2007; Schiff et al. Citation2012). Therefore, it is essential to understand the genetic and ecological diversity of siricid species, which can be crucial key factors for rapid responses to their damage on host trees.
In this study, some specimens of Tremex fuscicornis Fabricius, 1787 were collected from Pyeongchang-gun, Gangwon Province, South Korea (37°22′05.40″N, 128°23′39.60″E) in August 2019. These specimens were deposited in the Korea National Arboretum (KNA), Pocheon, South Korea (the number of the specimen used for the molecular analysis: 19CJ00496).
In this study, the complete mitochondrial genome of T. fuscicornis from Korea was sequenced using Illumina MiSeq (Macrogen, Inc., Seoul, South Korea). A total of 2,570,857,856 reads were analyzed to generate 8,541,056 base pairs of sequence and assembled in Geneious Prime (Kearse et al. Citation2012). The gene annotation was accomplished and circularity was checked using MITOS2 webserver (Bernt et al. Citation2013, http://mitos.bioinf.uni-leipzig.de/). The secondary structures of tRNA genes were analyzed by comparison with the nucleotide sequences of other insect tRNA sequences.
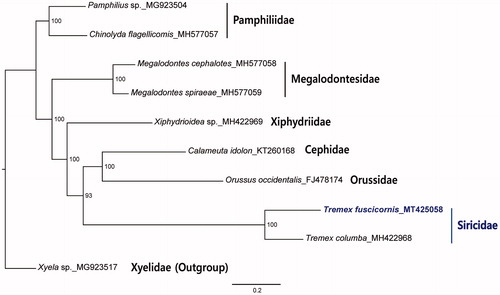
The maximum likelihood analysis of Siricidae was conducted based on concatenated 10 mitochondrial coding genes using RAxML version 8.1.2 (Stamatakis Citation2014) with 1000 bootstrap replications. Xyela sp. in Xyelidae was used as an outgroup. The analysis constructed a robust phylogenetic tree with high supports for all nodes (). The tree indicated that T. fuscicornis is a distinctive species in Siricidae, which is positioned as a sister group to (Cephidae + Orussidae) group. The mitogenome data of the species will be used for molecular-based species identification and phylogenetic studies of Siricoidea.
Figure 1. Phylogenetic tree based on the maximum-likelihood analysis of concatenated amino acid sequences of 10 mitochondrial protein-coding genes of T. fuscicornis and other Symphyta species using Geneious 11.0.5. The numbers beside the nodes indicate the percentages of 1000 bootstrap values. Xyela sp. was used as an outgroup. Alphanumeric terms indicate the GenBank accession numbers.
Disclosure statement
No potential conflict of interest was reported by the authors. The authors alone are responsible for the content and writing of the article.
Data availability statement
The data that support the findings of this study are openly available in [NCBI] at [https://www.ncbi.nlm.nih.gov/], reference number [MT425058].
Additional information
Funding
References
- Aguiar AP, Deans AR, Engel MS, Forshage M, Huber JT, Jennings JT, Johnson NF, Lelej AS, Longino JT, Lohrmann V, et al. 2013. Order Hymenoptera. Zootaxa. 3703(1):51–62.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo Metazoan Mitochondrial Genome Annotation Molecular Phylogenetics and Evolution. 69(2):313–319.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- National Institute of Biological Resources. 2019. National Species list of Korea. III. Insects (Hexapoda). Incheon (South Korea): National Institute of Biological Resources; p. 988.
- Pažoutová S, Šrůtka P. 2007. Symbiotic relationship between Cerrena unicolor and the horntail Tremex fuscicornis recorded in the Czech Republic. Czech Mycol. 59(1):83–90.
- Schiff NM, Goulet AH, Smith DR, Boudreault C, Wilson AD, Scheffler BE. 2012. Siricidae (Hymenoptera: Symphyta: Siricoidea) of the Western Hemisphere. Can J Arthropod Identif. 21:1–305.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Yan Y, Niu G, Zhang Y, Ren Q, Du S, Lan B, Wei M. 2019. Complete mitochondrial genome sequence of Labriocimbex sinicus, a new genus and new species of Cimbicidae (Hymenoptera) from China. PeerJ. 7:e7853.
