Abstract
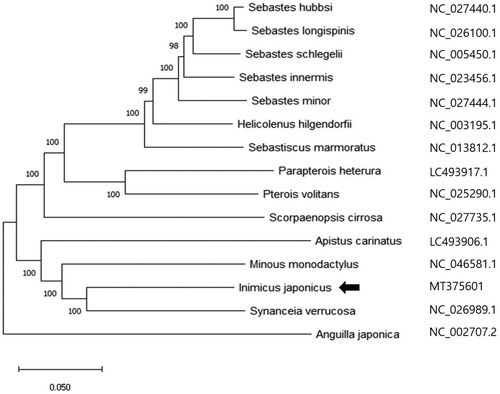
The complete mitochondrial genome (mitogenome) of the Inimicus japonicus was analyzed by next-generation sequencing in this study. The mitogenome is 16,978 base pairs (bp) long and codes for 22 transfer RNA (tRNA) genes, 13 protein-coding genes (PCGs), 2 ribosomal RNA genes (rRNA), and 1 non-coding control region. The overall nucleotide composition of the mitogenome is: 29.61% for A, 29.16% for T, 25.26% for C, and 15.97% for G. Twenty-two tRNAs range from 67 to 74 bp in length, and 2 rRNA (12S and 16S) were 953 and 1,687 bp long, respectively. Phylogenetic analysis by neighbor-joining (NJ) method indicated that I. japonicus showed considerable genetic similarity (82%), and had a closer relationship in the phylogenetic tree to Synanceia verrucosa.
Inimicus japonicus, common name: devil stinger, is a commercially important marine fish in East Asian countries. It widely inhabits the western Indo-Pacific region, including East China sea and Republic of Korea, at depths between 10 and 200 m (Nozaki et al. Citation2003; Wang et al. Citation2013). This species belongs to genus Inimicus, family Synanceiidae, and order Scorpaeniformes. Eleven species have been reported within the genus Inimicus (http://www.fishbase.org), but complete mitochondrial genome sequence has not been analyzed. In this study, we performed the first complete mitochondrial genome sequence analysis of I. japonicus using next-generation sequencing (NGS), and analyzed its phylogenetic relationship based on 14 mitogenome in the Scorpaenifomes order, with one available mitogenome in the Anguilliformes order as an outgroup.
The specimen of I. japonicus was collected from waters off the Seogwipo city, Jeju island, Republic of Korea (35°15′32.2″N, 126°40′58.3″E), and deposited in specimen room of Biotechnology Research Division, National Institute of Fisheries Science, Busan, Korea (Accession No. NFRDI-FI-TS-0056686). Total genomic DNA was extracted from the ventral fin, and the mitogenome was visualized in OrganellarGenomeDRAW v.1.3.1 (Greiner et al. Citation2019).
The assembled mitogenome of I. japonicus was 16,978 bp long and consisted of 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) (12S and 16S) genes, 13 protein-coding genes (PCGs), and a non-coding control region (D-Loop). The mitogenome was similar to other bony fish such as black sea bream Acanthopagrus schlegelii (Ma et al. Citation2016), Japanese amberjack Seriola quinqueradiata, Yellowtail amberjack Seriola lalandi (Oh and Jung Citation2019), and Longneck croaker Pseudotolithus typus (Kim et al. Citation2020). The mitochondrial genome sequences were deposited in the GenBank database (Genebank accession no. MT375601). The PCGs included 7 subunits of NADH dehydrogenase (ND), 3 from cytochrome c oxidase complex (COX), 1 from cytochrome b (Cytb), and the ATPase subunits 6 and 8 (ATP6 and ATP8). The overall nucleotide composition of the mitogenome was found to be as follows: 29.61% for A, 29.16% for T, 25.26% for C, and 15.97% for G. The A + T content (58.77%) was higher than the G + C content (41.23%). Most of the PCGs were initiated with the start codon ATG, except for COX1, which began with the GTG codon. Seven PCGs (COX1, ND1, ND4L, ND5, ND6, ATP6, and ATP8) ended with the stop codon TAA, and six PCGs (COX2, COX3, Cytb, ND2, ND3, and ND4) had an incomplete TA– or T–– stop codon. The 22 tRNA genes varies from 67 to 74 bp in length. The 12S rRNA was 953 bp long, located between the tRNA-Val and tRNA-Phe genes. The 16S rRNA was 1687 bp long, located between tRNA-Leu and tRNA-Val genes. The D-Loop was located between the tRNA-Phe and tRNA-Pro genes and was 1316 bp long.
The phylogenetic analysis of I. japonicus was performed using the complete mitochondrial DNA sequences from 14 species of Scorpaeniformes and 1 species of Anguilliformes using the neighbor-joining method (). Bootstrap values of 1000 replicates are shown beside each species. Our results show that I. japonicus has a closer relationship and shows considerable genetic similarity (82%) to Synanceia verrucosa. The results of this study will contribute toward the better understanding of family Synanceiidae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at [https://www.ncbi.nlm.nih.gov/genbank], accession number MT375601.
Additional information
Funding
References
- Greiner S, Lehwark P, Bock P. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–64.
- Kim J-O, Seo YB, Shin J, Yang J-Y, Kim G-D. 2020. Characterization of the complete mitochondrial genome of longneck croaker Pseudotolithus typus (Perciformes: Sciaenidae). Mitochondrial DNA Part B. 5(2):1167–1169.
- Ma X, Xie Z, Zhou L, Chen Y. 2016. The complete mitochondrial genome of Acanthopagrus schlegelii (Perciformes: Sparidae) with phylogenetic consideration. Mitochondrial DNA Part B. 1(1):150–151.
- Nozaki R, Takushima M, Mizuno K, Kadomura K, Yasumoto S, Soyano K. 2003. Reproductive cycle of devil stinger, Inimicus japonicus. Fish Physiol Biochem. 28(1–4):217–218.
- Oh DJ, Jung YH. 2019. Mitochondrial genome of Japanese amberjack, Seriola quinqueradiata, and yellowtail amberjack, Seriola lalandi. Mitochondrial DNA Part B. 4(1):826–827.
- Wang Y, Li L, Cui G, Lu W. 2013. Ontogenesis from embryo to juvenile and salinity tolerance of Japanese devil stinger Inimicus japonicus during early life stage. Springerplus. 2(1):289.