Abstract
In this study, we provide the report of a mitochondrial genome of Shaw’s sea snake (Hydrophis curtus). The total length of the mitogenome is 17839 bp, which was composed of 13 protein-coding genes (PCGs), 2 rRNA genes (12 s and 16 s rRNA), 22 tRNA genes. The base composition was 32.6% for A, 27.4% for T, 26.7% for C, and 13.3% for G. The sequence presented would be useful for future phylogenetic and evolutionary study of sea snakes.
The genus Hydrophis, also known as core Hydrophis group/lineage, belongs to the family Elapidae and containing 49 species (Sanders et al. Citation2013; Ukuwela et al. Citation2017). Shaw’s sea snake (Hydrophis curtus) is a viviparous species, belongs to the genus Hydrophis and possess several morphological adaptations that associate with their marine life, such as laterally compressed bodies, paddle-shaped tails and reduced ventral scales. A genomic research of Hydrophis curtus reveal the genetic mechanism of its marine adaptation (Peng et al. Citation2020). In our study, we obtained the mitochondrial sequence of Shaw’s sea-snake based on the published genome (Peng et al. Citation2020).
The Hydrophis curtus specimen was donated by the local fishermen from the coastal of Haikou, Hainan Province, China, and its DNA was stored in the Herpetological Museum of the Chengdu Institute of Biology, Chinese Academy of Sciences with the number 2160722004. The mitochondrial genome was generated from the raw data of Hydrophis curtus genome (PRJNA597425, library SRR10861676 was used) (Peng et al. Citation2020). To annotate the mitochondrial DNA sequence, a mitochondrial genome annotation web server (MITOS) was used to predicted the gene structure and then correct the mitogenomic structure manually (Bernt et al. Citation2013). The tRNA genes were scanned by tRNAScan-SE2.0 online website (http://lowelab.ucsc.edu/tRNAscan-SE/) (Lowe and Chan Citation2016), and the base composition was calculated by MEGA7 (Kumar et al. Citation2016).
The total length of the Hydrophis curtus mitochondrial genome is 17839 bp in size, with base compositions of 32.6% for A, 27.4% for T, 26.7% for C, and 13.3% for G. The mitogenome contained 13 PCGs, 2 rRNA genes (12 s and 16 s rRNA), 22 tRNA genes. For the 13 PCGs, the longest gene is ND5 (1770 bp), while the shortest gene is ATP8 (165 bp). The 22 tRNA genes varied in size from 57 to 73 bp of which the shortest one was tRNA-Ser and the longest was tRNA-Asn. The sequence length of 12 s and 16 s rRNA were 940 bp and 1473 bp, respectively, which were separated by tRNA-Val. Among these genes, 28 of 37 genes are encoded on the H-strand, containing 12 PCGs, 2 rRNA, 14 tRNA; and other 9 genes are located on the L-strand, containing ND6 and 8 tRNA. The mitogenome sequence together with gene annotations of Hydrophis curtus have been deposited in GenBank under the accession number MT712129.
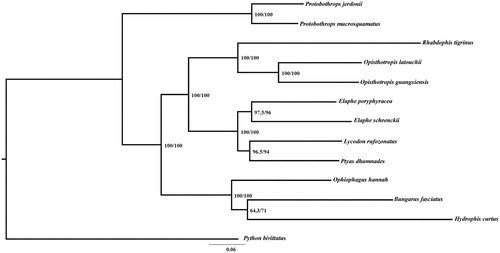
For convincing the mitochondrial DNA sequences, a maximum likehood (ML) phylogenetic tree () was re-constructed based on 12 H-strand coding genes using IQTREE (Lam-Tung et al. Citation2015). The sequence alignment was performed by MEGA7 (Kumar et al. Citation2016) with default settings, and a python script was written to concatenate the aligned sequences. Phylogenetic analysis showed that Hydrophis curtus was located within the lineage of Elapidae. We hope that this mitogenome announcement would provide useful resources for further phylogenetic and evolutionary studies.
Figure 1. A maximum likehood (ML) phylogenetic tree of 13 snakes. Numbers in parentheses are SH-aLRT support (%)/ultrafast bootstrap support (%). The species selected and corresponding GenBank accession number are shown as follows: Bungarus fasciatus (NC_011393), Elaphe poryphyracea (NC_012770), Elaphe schrenckii (NC_027605), Lycodon rufozonatus (NC_024559), Ophiophagus hannah (NC_011394), Opisthotropis latouchii (NC_046823), Protobothrops jerdonii (NC_021402), Protobothrops mucrosquamatus (NC_021412), Ptyas dhumnades (NC_028049), Python bivittatus (NC_021479), Rhabdophis tigrinus (NC_030210), Opisthotropis guangxiensis (MT_571495).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in “NCBI” at https://www.ncbi.nlm.nih.gov/, reference number MT712129.
References
- Bernt M, Donath A, J Uhling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lam-Tung N, Schmidt HA, Arndt VH, Quang MB. 2015. Iq-tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Peng CJ, Ren JL, Deng C, Jiang DC, Wang J, Qu JY, Chang J, Yan CC, Jiang K, Murphy RW, Wu DD, et al. 2020. The genome of Shaw’s sea snake (Hydrophis curtus) reveals secondary adaptation to its marine environment. Mol Biol Evol. 37(6):1744–1760.
- Sanders KL, Lee MS, Mumpuni, Bertozzi T, Rasmussen AR. 2013. Multilocus phylogeny and recent rapid radiation of the viviparous sea snakes (Elapidae: Hydrophiinae). Mol Phylogenet E. 66:575–591.
- Ukuwela KD, Lee MS, Rasmussen AR, De Silva A, Sanders KL. 2017. Biogeographic origins of the viviparous sea snake assemblage (Elapidae) of the Indian Ocean. Ceylon J Sci. 46(5):101.
