Abstract
Here, we report the complete chloroplast (cp) genome of Pinus densata. The complete chloroplast genome is 119,617 bp in length. There were 112 genes in the genome, including 73 protein-coding genes, 35 tRNA genes, and 4 rRNA genes. The overall GC was 38.5%, and the base of A, C, G, and T were 30.6, 19.3, 19.2, and 30.9%, respectively. Phylogenetic analysis showed that P. densata was relatively closely related to Pinus tabuliformis. These data may providing useful information for the phyletic evolution of P. densata within the Pinaceae family.
Pinus densata is an evergreen conifer tree species belonging to the Pinaceae family, is endemic to China. The species has a distinct niche, occurring exclusively in high-altitude regions ranging between 2700 and 4200 m in the southeastern Qinghai-Tibetan Plateau, China (Qiu et al. Citation2016). Genetic analyses suggest that P. densata originated from natural hybridization between Pinus tabuliformis and Pinus yunnanensis (Ma et al. Citation2006). Additionally, it is a kind of afforestation tree in western Sichuan and eastern Tibet that is associated with high economic value and ecological benefits. It is also an ecologically successful homoploid hybrid conifer associated with far-reaching evolutionary significance (Wang et al. Citation2011).
The chloroplast genome is a circular DNA molecule (Tonti-Filippini et al. Citation2017). It is highly conserved in plants and therefore ideal for ecological, evolutionary, and diversity studies (Wicke et al. Citation2011). To date, complete chloroplast genomes of several species from the Pinaceae family have been studied and deposited at the GenBank database, such as Pinus massoniana (Ni et al. Citation2020) and P. tabuliformis (Yu et al. Citation2017). However, few data are available regarding the P. densata cp genome. To gain the knowledge of its complete chloroplast genome, we determined the complete sequence of the chloroplast genome of P. densata. Additionally, we compared it with other known cp genomes to determine phylogenetic relationships among Pinaceae family.
The fresh leaves of P. densata were collected from Southwest Forestry University Kunming, China (Yunnan, China; geospatial coordinates: 102°45′41″E, 25°04′00″N). The voucher specimens of P. densata were deposited at the herbarium of Southwest Forestry University (accession number: SWFU-2020-GS02), and DNA samples were stored at the Key Laboratory for Forest Resources Conservation and Utilization in the Southwest Mountains of China Ministry of Education, Southwest Forestry University, Kunming, China. The total genomic DNA was extracted by using the Magnetic beads plant genomic DNA preps Kit (TSINGKE Biological Technology, Beijing, China). The genome skimming sequencing was conducted on the Illumina HiSeq 2000 Sequencing platform. Genome annotation was assembled with the program Geneious R8 (Biomatters Ltd, Auckland, New Zealand). Finally, the chloroplast DNA sequence with complete annotation information was submitted to the GenBank with the accession number MT732337.
The complete cp genome of P. densata is 119,617 bp in length, The overall GC content is 38.5%, the base of A, C, G and T were 30.6, 19.3, 19.2 and 30.9%, respectively. A total of 112 functional genes were contained in the cp genome, including 73 protein-coding genes, 35 tRNA genes, and 4 rRNA genes. Among them 12 genes (atpF, rpoC1, petB, petD, rpl16, rpl2, trnV-UAC, trnK-UUU, trnG-UCC, trnL-UAA, trnI-GAU and trnA-UGC) have single introns, while one gene (ycf3) has double introns.
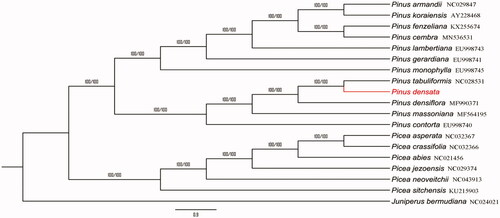
To explore the phylogenetic relationship of P. densata in the Pinaceae family, the phylogenetic tree was constructed based on the complete cp genome of P. densata and other 18 species. The result showed that P. densata appeared to be closely related to P. tabuliformis (). In conclusion, complete cp genome of P. densata is decoded for the first time, which will also provide essential DNA molecular data for further biological analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference number: MT732337.
Additional information
Funding
References
- Ma XF, Szmidt AE, Wang XR. 2006. Genetic structure and evolutionary history of a diploid hybrid pine Pinus densata inferred from the nucleotide variation at seven gene loci. Mol Biol Evol. 23(4):807–816.
- Ni Z, Ye YJu, Xu M, Xu L-A. 2020. Comparison among three methods for obtaining chloroplast genome sequences from the conifer Pinus massoniana. Genomics. 112(3):2459–2466.
- Qiu ZB, Yuan MM, Hai BZ, Wang L, Zhang L. 2016. Characterization and expression analysis of conserved miRNAs and their targets in Pinus densata. Biologia Plant. 60(3):427–434.
- Wang BS, Mao JF, Gao J, Zhao W, Wang XR. 2011. Colonization of the Tibetan Plateau by the homoploid hybrid pine Pinus densata. Mol Ecol. 20(18):3796–3811.
- Tonti-Filippini J, Nevill PG, Dixon K, Small L. 2017. What can we do with 1000 plastid genomes? Plant J. 90(4):808–818.
- Wicke S, Schneeweiss GM, Depamphilis CW, Muller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76(3-5):273–297.
- Yu ZD, Peng SB, Yang PH. 2017. The complete chloroplast genome of the southern Chinese pine Pinus tabuliformis (Pinales: Pinaceae). Mitochondrial DNA 28(1):13–14.