Abstract
The complete chloroplast genome sequence of Chaenomeles cathayensis and Chaenomeles thibetica was characterized from Illumina pair-end sequencing. The length of C. cathayensis and C. thibetica chloroplast genome of was 159,875 bp and 159,907 bp, respectively. The C. cathayensis chloroplast genome contained a large single-copy region (LSC) of 87,813bp, a small single-copy region (SSC) of 19,304 bp, and two inverted repeat (IR) regions of 26,379 bp, which is a quadripartite structure. Similarity, The C. thibetica chloroplast genome also contained a quadripartite structure, including a LSC region (87,851 bp), a SSC region (19,298 bp), and two IR regions (26,379 bp). The overall GC content of C. cathayensis and C. thibetica are both 36.57%. The C. cathayensis and C. thibetica chloroplast genome contains 130 complete genes, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The C. thibetica chloroplast genome contains 130 complete genes, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The neighbour-joining phylogenetic analysis showed that C. cathayensis and C. thibetica clustered together, indicated that C. cathayensis and C. thibetica have closed evolutionary relationship compared with other Chaenomeles species.
Introduction
The plant genus Chaenomeles Lindley is widely cultivated in East Asian, which have important ecologic and economic value (Phipps et al. Citation1990). Its fruit is rich in sugars, amino acids, flavonoids, saponins and organic acids (Rumpunen Citation2002; Ros et al. Citation2004). The fresh leaves of Chaenomeles cathayensis (Hemsley) C. K. Schneider and Chaenomeles thibetica T. T. Yu were respectively collected from Zhengan (28°35′50.61″N, 107°15′25.49″E) and Linzhi (30°17′34.29″N, 94°48′51.27″E) County, P. R. China. Fresh leaves were dried and stored in Herbarium of Research Institute of Subtropical Forestry of Chinese Academy of Forestry until DNA extraction (specimen Accession number SW190211 and SW190217). We extracted total genomic DNA from 25 mg dried leaf using a modified CTAB method (Doyle Citation1987). The whole-genome sequencing was then conducted by Illumina Hiseq 2000 platform (Illumina, San Diego, CA). We used SPAdes software (Bankevich et al. Citation2012) to assemble chloroplast genomes of C. cathayensis and C. thibetica. We annotated the chloroplast genome by prodigal v2.6.3 (Hyatt et al. Citation2010), hmmer v3.1(Prakash et al. Citation2017) and aragorn v1.2.38 (Laslett and Canback Citation2004).
The size of complete chloroplast genome of C. cathayensis (GenBank accession number MT561270 and MT561271) is 159,875 bp, containing a large single-copy region (LSC) of 87,813 bp, a small single-copy region (SSC) of 19,304 bp, and two inverted repeat (IR) regions of 26,379 bp. Similarity, The C. thibetica chloroplast genome also contained a quadripartite structure, including a LSC region (87,851 bp), a SSC region (19,298 bp), and two IR regions (26,379 bp). The overall GC content of C. cathayensis and C. thibetica are both 36.57%. The C. cathayensis and C. thibetica chloroplast genome contains 130 complete genes, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The C. thibetica chloroplast genome contains 130 complete genes, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes.
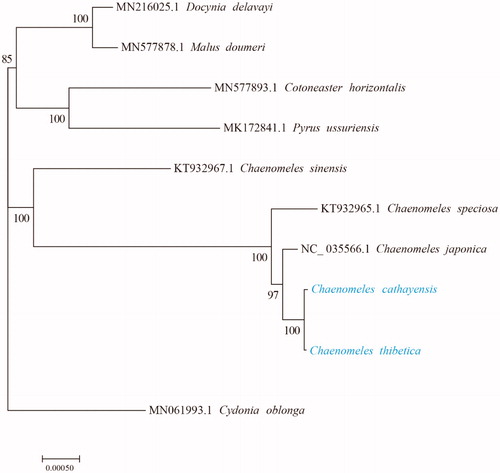
In order to explore the evolution relationship of C. cathayensis and C. thibetica, we used the complete chloroplast genomes sequence of C. cathayensis, C. thibetica, other Chaenomeles species, Docynia delavayi (Franchet) C. K. Schneider, Malus doumeri (Bois) A. Chevalier, Cotoneaster horizontalis Decaisne, Pyrus ussuriensis Maximowicz and Cydonia oblonga Miller to construct phylogenetic tree. Cydonia oblonga were considered as outgroup in this phylogenetic tree. All these 10 chloroplast genome sequences were aligned with MAFFT (Katoh and Standley Citation2013), and then the Maximum likelihood tree was constructed by RAxML v8.2.10 with GTR model, hill climbing algorithm and 100 bootstrap replicates (Stamatakis Citation2014). The results indicated that C. cathayensis and C. thibetica have closed evolutionary relationship compared with other Chaenomeles species (). In conclusion, all two complete chloroplast genomes of C. cathayensis and C. thibetica could provide important resource for the further studies on the evolution of Chaenomeles genus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, accession number MT561270 and MT561271.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 11:119.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16.
- Phipps JB, Robertson KR, Smith PG, Rohrer JR. 1990. A checklist of the subfamily Maloideae (Rosaceae). Can J Bot. 68(10):2209–2269.
- Prakash A, Jeffryes M, Bateman A, Finn RD. 2017. The HMMER web server for protein sequence similarity search. Curr Protoc Bioinformatics. 60(1):3.15.1–13.15.23.
- Ros J, Laencina J, Hellin P, Jordan M, Vila R, Rumpunen K. 2004. Characterization of juice in fruits of different Chaenomeles species. Food Science and Technology. 37(3):301–307.
- Rumpunen K. 2002. Chaenomeles: potential new fruit crop for Northern Europe. In: Janick J, Whipkey A, editors. Trends in new crops and new uses. Alexandria (VA): ASHS Press.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.