Abstract
The European map butterfly Araschnia levana (Linnaeus, 1758) is a species showing extreme seasonal polyphenism. The complete 15,207 bp circular A. levana mitogenome consisting of 81.6% AT nucleotides, was assembled by Illumina genome skimming. It includes 22 tRNAs, 13 protein-coding genes, 2 rRNAs, and a control region in the typical butterfly gene order. Araschnia levana COX1 features an atypical CGA start codon and ATP6, COX1, COX2, ND1, ND3, and ND4 have incomplete stop codons completed by 3′A residues added to the mRNA. Phylogenetic reconstruction places A. levana as a basal lineage within tribe Nymphalini, consistent with previous phylogenetic hypotheses.
The European map butterfly, Araschnia levana, is a lepidopteran that displays seasonal polyphenism, having distinct long-day (LD) and short-day (SD) morphologies based on the length of daylight exposure in the last larval instar (Vilcinskas and Vogel Citation2016). The LD morphology prevalent in the summer displays brown and white coloration, contrasted with the SD morphology prevalent in the springtime after overwintering which displays red and black coloration (Windig and Lammar Citation1999; Nijhout Citation2010). Due to the differences in morphologies of wing coloration and other physical characteristics, A. levana was once classified as two different species by Linnaeus (who originally described the SD form as Papilio levana and the LD form as P. prorsa (Linnaeus Citation1758)) but later it was recognized that this was a single species displaying extreme seasonal polyphenism (Hübner Citation1816; Koch and Bückmann Citation1987). Araschnia levana is widely distributed throughout Europe, and has spread into extreme northern and southern regions due to the effects of climate change (Parmesan et al. Citation1999). Here, we report the complete mitochondrial genome sequence of A. levana (GenBank MT712075) from specimen AL2017.1, collected in Embourg, Belgium (GPS 50.590N, 5.607E) in April 2017, that has been pinned, spread, and deposited in the Wallis Roughley Museum of Entomology, University of Manitoba (voucher WRME0507734).
DNA was prepared (McCullagh and Marcus Citation2015) and sequenced by Illumina NovaSeq6000 (San Diego, CA) (Marcus Citation2018). The mitogenome of A. levana was assembled by Geneious 10.0.9 from 9,941,511 paired 150 bp reads using a Mallika jacksoni (Lepidoptera: Nymphalidae) reference mitogenome (MT704828) (Alexiuk et al. CitationSubmitted) and was annotated with respect to sequences from M. jacksoni and Junonia stygia (MN623383) (Living Prairie Mitogenomics Consortium Citation2020). The A. levana nuclear rRNA repeat (GenBank MT750296) was also assembled and annotated using the rRNA repeat from M. jacksoni (MT704831) as a reference sequence.
The A. levana circular 15,207 mitogenome assembly was composed of 5891 paired reads with nucleotide composition: 40.5% A, 10.9% C, 7.4% G, and 41.2% T. The gene composition and order in A. levana is identical to all known nymphalid mitogenomes (Linard et al. Citation2017). Araschnia levana COX1 features an atypical CGA start codon as in many other insects (Liao et al. Citation2010). Araschnia levana ND2 has an ATA start codon, which is used infrequently in insect mitochondria, but is fairly common in the mitochondria of some other animal groups (Okimoto et al. Citation1990; Han et al. Citation2016). The mitogenome contains two protein-coding genes (COX1, COX2) with single-nucleotide (T) stop codons, and four protein-coding genes (ATP6, ND1, ND3, ND4) with two-nucleotide (TA) stop codons completed by post-transcriptional addition of 3′A residues. The locations and structures of tRNAs were determined using ARWEN v.1.2 (Laslett and Canback Citation2008). tRNAs have typical cloverleaf secondary structures except for trnS (AGN) where the dihydrouridine arm is replaced by a loop, while the mitochondrial rRNAs and control region are typical for Lepidoptera (McCullagh and Marcus Citation2015).
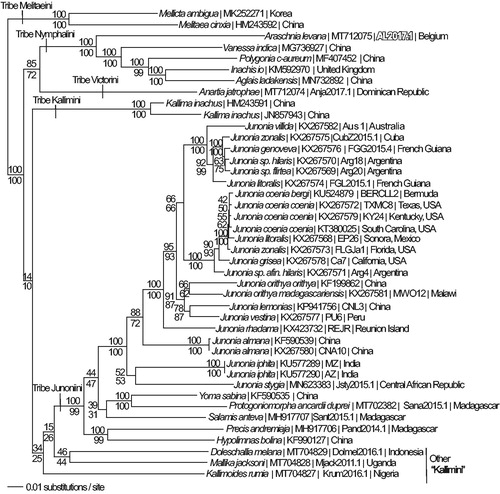
We reconstructed a phylogeny using complete mitogenomes from A. levana and 41 additional mitogenomes from subfamily Nymphalinae (Lalonde and Marcus Citation2019a, Citation2019b; Chen et al. Citation2020; Alexiuk et al. CitationSubmitted; Hamilton et al. CitationSubmitted; Lalonde and Marcus CitationSubmitted; Payment et al. CitationSubmitted-a, Submitted-b). Mitogenome sequences were aligned in CLUSTAL Omega (Sievers et al. Citation2011) and analyzed by parsimony and maximum likelihood (model selected by jModeltest 2.1.7 (Darriba et al. Citation2012) and likelihood ratio test (Huelsenbeck and Rannala Citation1997) in PAUP* 4.0b8/4.0d78 (Swofford Citation2002) (). Phylogenetic analysis places the A. levana mitogenome as the basal lineage among the available sequenced mitogenomes from the tribe Nymphalini, which is consistent with previous molecular phylogenetic studies of family Nymphalidae (Wahlberg et al. Citation2005, Wahlberg et al. Citation2009).
Figure 1. Maximum likelihood phylogeny (GTR + G model, G = 0.2330, likelihood score 117762.66543) of Araschnia levana (Tribe Nymphalini), 4 additional mitogenomes from tribe Nymphalini, 29 mitogenomes from tribe Junonini, 5 from Kallimini, 1 from Victorini, and 2 outgroup from tribe Melitaeini in subfamily Nymphalinae based on 1 million random addition heuristic search replicates (with tree bisection and reconnection). One million maximum parsimony heuristic search replicates produced 16 trees (parsimony score 20,698 steps) which differ from one another only by the arrangement of Junonia coenia mitogenomes and one of which has an identical tree topology to the maximum likelihood tree depicted here. Numbers above each node are maximum likelihood bootstrap values and numbers below each node are maximum parsimony bootstrap values (each from 1 million random fast addition search replicates).
Acknowledgements
We thank Rayna Hamilton and Josephine Payment for their constructive criticism on this manuscript and Genome Quebec for assistance with library preparation and sequencing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference numbers MT712075 and MT750296.
Additional information
Funding
References
- Alexiuk MR, Marcus JM, Lalonde MML. Submitted. The complete mitochondrial genome of the Jackson’s Leaf butterfly Mallika jacksoni (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA Part B: Resources Manuscript # TMDN-2020-1201, 1st round of review. Submitted 13 July 2020.
- Chen K, Si C, Pan Z, Hao J. 2020. The complete mitochondrial genome of Aglais ladakensis (Lepidoptera: Nymphalidae: Nymphalinae. Mitochondrial DNA Part B. 5(1):639–641.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Hamilton RV, Marcus JM, Lalonde MML. Submitted 2020 Jul. The complete mitochondrial genome of the black dead leaf butterfly Doleschallia melana (Insecta: Lepidoptera: Nymphalidae) Mitochondrial DNA B Resour. - Manuscript # TMDN-2020-1200, Accepted 19 August 2020.
- Han Z, Wang G, Xue T, Chen Y, Li J. 2016. The F-type complete mitochondrial genome of Chinese freshwater mussels Cuneopsis pisciculus. Mitochondrial DNA Part A:3376–3377.
- Hübner J. 1816. Verzeichnis bekannter Schmetterlinge. Augsburg: bey dem Verfasser zu Finden.
- Huelsenbeck JP, Rannala B. 1997. Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science. 276(5310):227–232.
- Koch PB, Bückmann D. 1987. Hormonal control of seasonal morphs by the timing of ecdysteroid release in Araschnia levana L. (Nymphalidae: Lepidoptera). J Insect Physiol. 33(11):823–829.
- Lalonde MML, Marcus JM. 2019a. The complete mitochondrial genome of the Madagascar banded commodore butterfly Precis andremiaja (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA Part B. 4(1):277–279.
- Lalonde MML, Marcus JM. 2019b. The complete mitochondrial genome of the Madagascar mother-of-pearl butterfly Salamis anteva (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA Part B. 4(1):296–298.
- Lalonde MML, Marcus JM. Submitted 2020 Jul. The complete mitochondrial genome of the Malagasy clouded mother-of-pearl butterfly Protogoniamorpha ancardii duprei (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA Part B Resources. doi: 10.1080/23802359.2020.1810156.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Liao F, Wang L, Wu S, Li Y-P, Zhao L, Huang G-M, Niu C-J, Liu Y-Q, Li M-G. 2010. The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int J Biol Sci. 6(2):172–186.
- Linard B, Arribas P, Andujar C, Crampton-Platt A, Vogler AP. 2017. The mitogenome of Hydropsyche pellucidula (Hydropsychidae): first gene arrangement in the insect order Trichoptera. Mitochondrial DNA Part A. 28(1):71–72.
- Linnaeus. 1758. Systema Naturae per Regna Tria Naturae, Secundum Clases, Ordines, Genera, Species, cum Characteribus, Differentiis, Symonymis, Locis. Tomis I. 10th ed. Syst Nat. 1:473.
- Living Prairie Mitogenomics Consortium. 2020. The complete mitochondrial genome of the brown pansy butterfly, Junonia stygia (Aurivillius, 1894), (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA Part B. 5:41–43.
- Marcus, JM 2018. Our love-hate relationship with DNA barcodes, the Y2K problem, and the search for next generation barcodes. AIMS Genetics. 5 (1): 1–23.
- McCullagh BS, Marcus JM. 2015. The complete mitochondrional genome of Lemon Pansy, Junonia lemonias (Lepidoptera: Nymphalidae: Nymphalinae). J Asia Pac Entomol. 18(4):749–755.
- Nijhout HF. 2010. Molecular and physiological basis of color pattern formation. Adv Insect Physiol. 38:219–265.
- Okimoto R, Macfarlane JL, Wolstenholme DR. 1990. Evidence for the frequent use of TTG as the translation initiation codon of mitochondrial protein genes in the nematodes, Ascaris suum and Caenorhabditis elegans. Nucleic Acids Res. 18(20):6113–6118.
- Parmesan C, Ryrholm N, Stefanescu C, Hill JK, Thomas CD, Descimon H, Huntley B, Kaila L, Kullberg J, Tammaru T, et al. 1999. Poleward shifts in geographical ranges of butterfly species associated with regional warming. Nature. 399(6736):579–583.
- Payment JE, Marcus JM, Lalonde MML. Submitted-a 2020 Jun. The complete mitochondrial genome of the African leaf butterfly Kallimoides rumia (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA Part B: Resources Manucript # TMDN-2020-1199.R1, 2nd round of review. Resubmitted 14 August 2020.
- Payment JE, Marcus JM, Lalonde MML. Submitted-b 2020 Jul. Phylogenetic analysis of the complete mitochondrial genome of the white peacock butterfly Anartia jatrophae saturata (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA Part B: Resources Manucript # TMDN-2020-1181, 2nd round of review. Resubmitted 20 August 2020.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.
- Swofford DL. 2002. PAUP*. Phylogenetic analysis using Parsimony (*and other methods). Version 4. Sunderland (MA): Sinauer Associates.
- Vilcinskas A, Vogel H. 2016. Seasonal phenotype-specific transcriptional reprogramming during metamorphosis in the European map butterfly Araschnia levana. Ecol Evol. 6(11):3476–3485.
- Wahlberg N, Brower AVZ, Nylin S. 2005. Phylogenetic relationships and historical biogeography of tribes and genera in the subfamily Nymphalinae (Lepidoptera: Nymphalidae). Biol J Linn Soc. 86(2):227–251.
- Wahlberg N, Leneveu J, Kodandaramaiah U, Peña C, Nylin S, Freitas AVL, Brower AVZ. 2009. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. Proc Biol Sci. 276(1677):4295–4302.
- Windig JJ, Lammar P. 1999. Evolutionary genetics of seasonal polyphenism in the map butterfly Araschnia levana (Nymphalidae: Lepidoptera). Evol Ecol Res. 1:875–894.
