Abstract
The increasing interest in understanding the evolutionary relationship between members of the Pteropodidae family has been greatly aided by genomic data from the Old World fruit bats. Here we present the complete mitogenome of Geoffroy’s rousette, Rousettus amplexicaudatus found in Peninsular Malaysia . The mitogenome constructed is 16,511bp in length containing 37 genes; 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and a D-loop region. The overall base composition is estimated to be 32.28% for A, 25.64% for T, 14.09% for G and 27.98% for C, indicating a slightly AT rich feature (57.93%). A phylogenetic and BLASTn analysis against other available mitogenomes showed Malaysian R. amplexicaudatus matched 98% similarity to the same species in Cambodia and Vietnam. However, it differed considerably (92.53% similarity) with the same species in the Philippines. This suggests flexibility in Rousettus sp. with regards to adapting to mesic and dry habitats, ability for long-distance dispersal and remarkably precise lingual echolocation thus supporting its wide-range distribution and colonization. Further taxonomical and mitogenomic comparatives are required in resolving the evolutionary relationship between Rousettus spp.
In tropical and subtropical regions of the Old World, fruit bats are an important seed dispersal agents that provide vital ecological services (Chan et al. Citation2020). Amidst fruit bats, the echolocating bats of the genus Rousettus in the Pteropodidae family are widely distributed from Asia to Africa (Hassanin et al. Citation2020). There are ten Rousettus sp. reported worldwide and six of these species occurring within Southeast Asia (Francis Citation2019). Rousettus spp. are selected as the best candidate amongst the Pteropodidae family to study evolutionary relationships as it is the only genus that has the capability of long-distance dispersal and has showed remarkable ecological flexibility in characteristics such as a well-developed echolocation system (Almeida et al. Citation2016; Stribna et al. Citation2019; Hassanin et al. Citation2020). However, lack of genomic data of Asian Rousettus sp. could hinder the extensive study of the genus. In this study, we sequenced and provided the whole mitochondrial genome of R. amplexicaudatus, which is the first mitogenome of Rousettus species from Malaysia.
Muscle tissue was obtained from Gading Forest Reserve, Selangor, Malaysia (Latitude: 3° 21′ 0″ N Longitude: 101° 15′ 0″ E) (Munian et al. Citation2020). The specimen voucher number MZF1072 was deposited in the Zoological Collection of Forest Research Institute Malaysia (FRIM). The methodologies used for DNA isolation, library construction, read assembly, and gene annotation are described in (Mohd Salleh et al. Citation2017; Jahari et al. Citation2020). The mitogenome of R. amplexicaudatus from this study (MT259592) is a circular molecule with 16,511 bp in length. Similar to other volant mammals, it contained 13 protein-coding genes (PCGs), 22 transfer RNA genes, 2 ribosomal RNA genes and 1 D-loop region (Yoon et al. Citation2016; Hassanin et al. Citation2020). The overall base composition of R. amplexicaudatus is estimated to be 32.28% for A, 25.64% for T, 14.09% for G and 27.98% for C, indicating a slightly AT rich feature (57.93%). The total length of the protein-coding gene sequences (PCGs) is 11,405bp. The total length of the 22 tRNA genes is 1509 bp, ranging from 57 bp (tRNASer) to 74 bp (tRNALeu). The 12S rRNA gene length is 969 bp and the 16S rRNA gene length is 1571 bp, and are located between the tRNAPhe and tRNALeu, and are separated by the tRNAVal gene. The control region is located between tRNAPro and tRNAPhe genes. The genes mostly located on the heavy (H) strand except for NAD6 and eight tRNAs genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, tRNAPro), which were found to be located on the lower (L) strand.
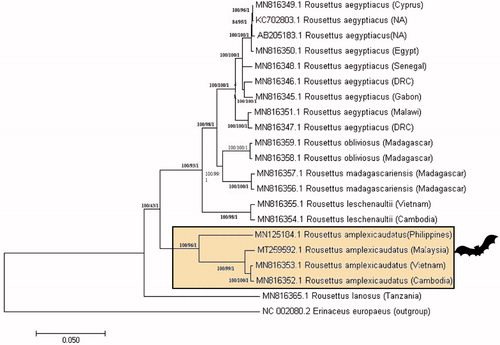
The Malaysian R. amplexicaudatus mitogenome shows 98% similarity to the same species from Cambodia (MN816352.1) and Vietnam (MN816353.1) (Hassanin et al. Citation2020) available in Genbank. Interestingly, it diverged by nearly 8% (92.53% match) for the same species originating from the Philippines (MN125184.1) (Mendoza & Fontanilla Citation2019). In , the phylogenetic analysis of this relationship of the Rousettus sp. mitogenomes is depicted. The phylogenetic tree generated a monophyletic clade of R. amplexicaudatus from Malaysia, Cambodia and Vietnam meanwhile; Philippines R. amplexicaudatus formed another branch. Unlike other fruit bats, Rousettus sp. are reported to be more adaptive to certain biogeographic barriers. Their flexibility to adapting in mesic and dry habitats, ability for long-distance dispersal and remarkably precise lingual echolocation support the species wide-range distribution and colonization (Happold & Happold Citation2013; Stribna et al. Citation2019; Hassanin et al. Citation2020). Therefore, further taxonomical and mitogenome comparative studies are essential to resolve the evolutionary relationships of this widely distributed Rousettus genus especially within Southeast Asia region.
Figure 1. Phylogenetic tree constructed using the whole mitogenome of Malaysian R. amplexicaudatus (MT259592) and other Rousettus sp. from different countries. The tree was generated from NJ/ML/Bayesian method using hedgehog as an outgroup. Bootstrap values generated from 1000 replicates for NJ/ML/Bayesian analysis. The number at each node indicated the bootstrap probability of NJ/ML/Bayesian analysis. (NA: not available; DRC: Democratic Republic of the Congo).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Data availability statement
The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, accession number MT259592.
Additional information
Funding
References
- Almeida FC, Giannini NP, Simmons NB. 2016. The evolutionary history of the African fruit bats (Chiroptera: Pteropodidae). Acta Chiropterologica/Museum and Institute of Zoology. Pol Acad Sci. 18(1):73–108.
- Chan AAQ, Aziz SA, Clare EL, Coleman JL. 2020. Diet, ecological role and potential ecosystem services of the fruit bat, Cynopterus brachyotis, in a tropical city. Urban Ecosyst. 23(1):1–13.
- Francis C. 2019. Field guide to the mammals of South-east Asia. 2nd ed. London: Bloomsbury Publishing.
- Happold M, Happold DC. 2013. Mammals of Africa: hedgehogs, shrews and bats. London: Bloomsbury Pub.
- Hassanin A, Bonillo C, Tshikung D, Pongombo Shongo C, Pourrut X, Kadjo B, Nakouné E, Tu VT, Prié V, Goodman SM. 2020. Phylogeny of African fruit bats (Chiroptera, Pteropodidae) based on complete mitochondrial genomes. J Zool Syst Evol Res. 39:1.
- Jahari PNS, Abdul Malik NF, Shamsir MS, Gilbert MTP, Mohd Salleh F. 2020. The first complete mitochondrial genome data of Hippocampus kuda originating from Malaysia. Data Brief. 31:105721.
- Mendoza RVD, Fontanilla IKC. 2019. Whole mitochondrial genome of a Geoffroy’s Rousette, Rousettus amplexicaudatus (Pteropodidae). Mitochondrial DNA Part B. 4(2):3546–3548.
- Mohd Salleh F, Ramos-Madrigal J, Peñaloza F, Liu S, Mikkel-Holger SS, Riddhi PP, Martins R, Lenz D, Fickel J, Roos C, et al. 2017. An expanded mammal mitogenome dataset from Southeast Asia. Gigascience. 6(8):1–8.
- Munian K, Azman SM, Ruzman NA, Fauzi NFM, Zakaria AN. 2020. Diversity and composition of volant and non-volant small mammals in northern Selangor State Park and adjacent forest of Peninsular Malaysia. BDJ. 8:e50304.
- Stribna T, Romportl D, Demjanovič J, Vogeler A, Tschapka M, Benda P, Horáček I, Juste J, Goodman SM, Hulva P. 2019. Pan African phylogeography and palaeodistribution of rousettine fruit bats: ecogeographic correlation with Pleistocene climate vegetation cycles. J Biogeogr. 46(10):2336–2349.
- Yoon KB, Kim JY, Park YC. 2016. Characteristics of complete mitogenome of the lesser short-nosed fruit bat Cynopterus brachyotis (Chiroptera: Pteropodidae) in Malaysia. Mitochondrial DNA A DNA Mapp Seq Anal. 27(3):2091–2092.
