Abstract
Here, we present the mitogenome of the blood feeding leech Haementeria acuecueyetzin (Hirudinida: Glossiphoniidae) based on specimens collected in Tabasco, Mexico. The circular genome is 14,985 bp in length, and consists of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and an AT-rich control region. Phylogenetic analysis based on the 13 protein-coding genes and two rRNA genes places H. acuecueyetzin sister to H. officinalis within the family Glossiphoniidae. Mitochondrial gene order in H. acuecueyetzin is consistent with other members of Clitellata with no evidence of gene gain/loss, duplication, or rearrangement.
Haementeria acuecueyetzin Oceguera-Figueroa (Citation2008) is a blood-feeding leech that inhabits large parts of southern Mexico. It feeds on a variety of vertebrates including cattle, manatees, crocodiles, and even humans (Oceguera-Figueroa Citation2008; Pérez-Flores et al. Citation2016; Charruau et al. Citation2020). The genus includes 13 species geographically restricted to the Neotropics (Oceguera-Figueroa Citation2012; Oceguera-Figueroa and León-Règagnon Citation2014). Haementeria species have been used as model organisms in studies of neurophysiology (De-Miguel et al. Citation2001), development (Sawyer et al. Citation1981), and bacterial symbioses (Manzano-Marín et al. Citation2015), among other areas.
On 18 March 2018, a total of 30 specimens of H. acuecueyetzin were collected in Tabasco, Mexico (17° 40′ 56.53″N, 93° 00′07.92″W). For a parallel project regarding the leech bacterial endosymbionts, total DNA of a mixed pool of 120 bacteria bearing organs or bacteriomes from 30 leeches (four from each individual) was extracted using a commercial extraction kit (DNeasy Blood & Tissue Kit, Qiagen, Hilden, Germany). DNA libraries were constructed using the NGS Nextera FLEX DNA library preparation kit (Illumina Inc., San Diego, CA) according to the manufacturer’s protocol and total DNA was multiplexed together with 11 other samples and sequenced on a single lane on the HiSeqX platform (150 bp paired-end) at SickKids, Toronto, Canada. Adapters were trimmed with the FastX-Toolkit (Gordon and Hannon Citation2010), and sequencing quality was assessed with FastQC v0.11.9 (Andrews Citation2019). Reads were assembled using SPAdes v3.13.0 (Bankevich et al. Citation2012) and, subsequently, taxonomic assignment of each contig was performed by a local BLASTx search (Altschul et al. Citation1997) against the proteomes of Providencia siddallii (CVRF01000001–CVRF01000004), Helobdella robusta (AF178680) and Haementeria officinalis (LT159848) as reference for bacterial endosymbiont, host (leech), and leech mitochondrial sequences, respectively. We recovered a linear contig of ∼15 Kbp with strong similarity in both nucleotide identity and length when compared to Haementeria officinalis, the only previously sequenced mitogenome from a congener. Online BLASTn and BLASTx searches confirmed the leech affinity of the contig and its components (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Shorter sequencing reads were mapped to the single contig using Bowtie 2.3.5.1 (Langmead and Salzberg Citation2012) and reassembled, resulting in a sequence of >15 Kbp in length and a coverage of 1085x. Low complexity regions at both 5′- and 3′-ends were trimmed for a final sequence of 14,985 bp in length. The resulting sequence was annotated using the MITOS web server (Bernt et al. Citation2013) with one round of manual curation to correctly define 5′- and 3′-ends of genes, resulting in a complete mitogenome consisting of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and an AT-rich control region. The newly reported mitogenome was deposited in GenBank under accession number MT683771. Voucher specimens for H. acuecueyetzin are deposited in the Colección Nacional de Helmintos, Universidad Nacional Autónoma de México, Mexico City, Mexico (CNHE 11386).
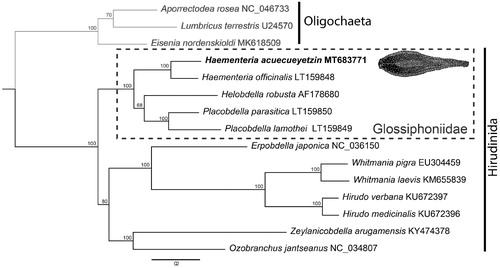
Sequences of the 13 protein coding genes and two ribosomal RNAs were used to infer the phylogenetic position of H. acuecueyetzin within Hirudinea using all available mitogenomes from GenBank. A maximum-likelihood tree was obtained using RaxML v.8.2.4 (Stamatakis Citation2006), employing the model GTRGAMMAI and setting bootstrap replicates as autoMRE function. Phylogenetic analysis places H. acuecueyetzin as the sister taxon to H. officinalis, within a clade exclusively formed by other members of Glossiphoniidae. The glossiphoniid clade, in turn, is the sister to the rest of hirudineans (). Mitochondrial gene order in H. acuecueyetzin is consistent with that of other members of Clitellata with no evidence of gene gain/loss, duplication, or rearrangement.
Figure 1. Phylogenetic hypothesis inferred by maximum-likelihood using 13 protein-coding genes and two ribosomal genes sourced from all available leech mitogenomes on GenBank. Haementeria acuecueyetzin places as the sister taxon to H. officinalis within a larger clade formed by members of Glossiphoniidae (dotted box). The ingroup (Hirudinida) is represented by black branches and Oligochaeta (outgroup) in gray branches.
Acknowledgements
We thank Misael Garrido-Casas for the illustration of Haementeria acuecueyetzin.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/genbank/, reference number MT683771.
Additional information
Funding
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402.
- Andrews S. 2019. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Charruau P, Oceguera-Figueroa A, Cedeño-Vázquez JR, Pérez-Rivera SD. 2020. Record of leech parasitism in Crocodylus moreletii (Duméril & Bibron, 1851) from Mexico. Comp Parasitol. 87(1):89–92.
- De-Miguel FF, Vargas-Caballero M, García-Pérez E. 2001. Spread of synaptic potentials through electrical synapses in Retzius neurons of the leech. J Exp Biol. 204(Pt 19):3241–3250.
- Gordon A, Hannon GJ. 2010. Fastx-toolkit. FASTQ/A short-reads pre-processing tools. Unpublished. http://hannonlab.cshl.edu/fastx_toolkit/.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Manzano-Marín A, Oceguera-Figueroa A, Latorre A, Jiménez-García LF, Moya A. 2015. Solving a bloody mess: B-vitamin independent metabolic convergence among gammaproteobacterial obligate endosymbionts from blood-feeding arthropods and the leech Haementeria officinalis. Genome Biol Evol. 7(10):2871–2884.
- Oceguera-Figueroa A. 2008. A new glossiphoniid leech from Catemaco Lake, Veracruz, México. J Parasitol. 94(2):375–380.
- Oceguera-Figueroa A. 2012. Molecular phylogeny of the New World bloodfeeding leeches of the genus Haementeria and reconsideration of the biannulate genus Oligobdella. Mol Phylogenet Evol. 62(1):508–514.
- Oceguera-Figueroa A, León-Règagnon V. 2014. Biodiversidad de sanguijuelas (Annelida: Euhirudinea) en México. Rev Mex Biodivers. 85:183–189.
- Pérez-Flores J, Rueda-Calderon H, Kvist S, Siddall ME, Oceguera-Figueroa A. 2016. From the Worm in a Bottle of Mezcal: iDNA confirmation of a leech parasitizing the Antillean Manatee. J Parasitol. 102(5):553–555.
- Sawyer RT, Lepont F, Stuart DK, Kramer AP. 1981. Growth and reproduction of the giant glossiphoniid leech Haementeria ghilianii. Biol Bull. 160(2):322–331.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
