Abstract
The snow petrel, Pagodroma nivea is a small sea bird endemic to Antarctica and the Southern Ocean. Herein, we assembled a complete mitochondrial genome of the snow petrel as a first revealed genetic resource in Pagodroma family. The mitogenome is 17,279 bp in length and consists of 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes. Base composition is 31.3% A, 25.3% T, 30.1% C, and 13.2% G with CG content of 43.4%. These results will provide a useful basis for further genetic and phylogenetic studies of this species.
The snow petrel (Pagodroma nivea Forster, G, 1777) is a small sea bird endemic to Antarctica and the Southern Ocean (Croxall et al. Citation1995). The species is a member of the family Procellariidae, order Procellariiformes, and is the only species representing the genus Pagodroma. It is divided into two subspecies (P. nivea and P. major) but taxonomic status remains controversial (Carrea et al. Citation2019). Snow petrels exhibit broad geographic distribution and a large, relatively stable global population (Birdlife International Citation2020). Despite this, few genetic studies have been conducted (Croxall et al. Citation2012). Also, mitochondrial genome was not reported in Pagodroma family until now. Thus, elucidation of the snow petrel mitochondrial genome will provide a basis for improved understanding of their phylogeny and evolution.
In this study, we sequenced and analyzed the complete mitogenome of P. nivea (GenBank: MT726204). A single specimen (proof number SP1) was collected in Bransfield Strait, Antarctica (62°25′52.8′′S, 58°42′20.2′′W), on 12 April 2018 and stored at the Korea Polar Research Institute (KOPRI), Incheon, South Korea. Total genomic DNA was extracted from muscle tissue using a DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany). An Illumina paired-end (PE) library was prepared according to the manufacturer’s instructions, and sequencing was performed using an Illumina sequencing platform by commercial company (Phyzen, Seongnam, South Korea). High-quality PE reads of about 1.03 Gb were obtained after trimming and were de novo assembled using CLC genome assembler version 4.21 (CLC Inc., Aarhus, Denmark). From the initially assembled contigs, those with mitogenome-derived sequences were further processed to generate a single draft sequence, as previously described (Lee et al. Citation2018). The draft sequence was manually corrected and gap-filled via a series of PE read mappings. The final complete mitogenome sequence was annotated using GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq-app.html) and manually curated using the Artemis annotation tool (Rutherford et al. Citation2000) with NCBI BLASTN searches (https://blast.ncbi.nlm.nih.gov). A maximum likelihood (ML) phylogenetic tree was constructed: multiple alignment of 13 protein-coding gene (PCG) sequences in mitochondrial genomes were performed using MAFFT (Katoh and Standley Citation2013) and used Tamura-Nei model and 1000 bootstrapping options via MEGA version 7.0 program (Pennsylvania State University, USA) (Kumar et al. Citation2016).
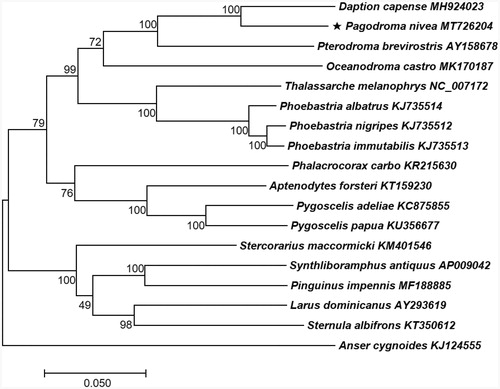
The resulting complete P. nivea mitochondrial genome sequence is 17,279 bp in length, including 13 PCGs, 22 tRNA genes, and two rRNA genes. Nucleotide composition is as follows: 31.3% A, 25.3% T, 30.1% C, and 13.2% G with a 43.4% GC content. Phylogenetic relationship revealed that P. nivea and Daption capense are a sister species and Pterodroma brevirostris and Oceanodroma castro belong to the same lineage ().
Acknowledgments
We appreciate the invaluable assistance in sample collection provided by Yeontae Gim.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the finding of this study are openly available in NCBI at (https://www.ncbi.nlm.nih.gov), reference number [MT726204] and available from the corresponding author.
Additional information
Funding
References
- BirdLife International. 2020. Species factsheet: Pagodroma nivea. [accessed 2020 Jul 03]. http://www.birdlife.org.
- Carrea C, Burridge CP, Wienecke B, Emmerson LM, White D, Miller KJ. 2019. High vagility facilitates population persistence and expansion prior to the Last Glacial Maximum in an Antarctic top predator: the snow petrel (Pagodroma nivea). J Biogeogr. 46(2):442–453.
- Croxall JP, Butchart SHM, Lascelles B, Stattersfield AJ, Sullivan B, Symes A, Taylor P. 2012. Seabird conservation status, threats and priority actions: a global assessment. Bird Conserv Int. 22(1):1–34.
- Croxall JP, Steele WK, Mcinnes SJ, Prince PA. 1995. Breeding distribution of the snow petrel Pagodroma nivea. Marine Ornithol. 23:69–100.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lee HO, Choi JW, Baek JH, Oh JH, Lee SC, Kim CK. 2018. Assembly of the mitochondrial genome in the campanulaceae family using Illumina low-coverage sequencing. Genes. 9(8):383.
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics. 16(10):944–945.