Abstract
Ichang papeda (Citrus cavaleriei) is an endemic perennial plant in China. In this study, we assembled and annotated the complete chloroplast genome of Yuanjiang wild Ichang papeda using Illumina Hiseq-2500 sequencing data. The chloroplast genome is constituted of 160,996 bp, containing a 87,634 bp large single-copy region, a 18,762 bp small single-copy region, and a pair of 27,300 bp inverted repeat regions. The chloroplast genome contains 114 unique genes, including 80 protein-coding genes, 30 tRNAs and 4 rRNAs. Phylogenetic analysis showed that the relationship between the chloroplast gennomes of C. cavaleriei and C. reticulata is the closest, which consistently support their chloroplast relationships.
Ichang papeda (Citrus cavaleriei, once named as Citrus ichangensis), an original Citrus species with large wing leaves, is firstly discovered in Xingshan county, Hubei province of China (Chen et al. Citation2012; Yang et al. Citation2017). Nowadays, its distributions in Yunnan, Hunan, Guangxi, Guizhou, Sichuan, Chongqing and Shanxi provinces of China have also been reported. The Yunnan province of China, an important Citrus original center, is rich of endemic and wild Citrus species (Wu et al. Citation2018). And the wild Ichang papeda population in Yuanjiang county of Yunnan province is recognized the largest and the oldest (Chen et al. Citation2012). Current researches on Yuanjiang wild Ichang papeda were mainly focused on its genetic diversity and evolution from aspects of phenotype, stress resistance, molecular markers and so on (Chen et al. Citation2012; Yang et al. Citation2017). In the present study, to provide genetic information of Yuanjiang wild Ichang papeda, we assembled and annotated the complete chloroplast genome, and investigated its phylogenetic relationships with other Citrus species.
The specimen of Yuanjiang wild Ichang papeda was isolated from Mountain Nizubai, Yicibei village, Wadie town, Yuanjiang county, Yunnan province, China (23°29′33.036″N; 102°17′9.276″E) and samples were deposited at the Institute of Tropical and Subtropical Cash Crops, Yunnan Academy of Agricultural Sciences. The leaf genomic DNA of Yuanjiang wild Ichang papeda was extracted using the CTAB method (Xie et al. Citation2020) and stored at the Institute of Tropical and Subtropical Cash Crops, Yunnan Academy of Agricultural Sciences (No. YJYCC01). The whole genomic DNA re-sequencing was performed on the Illumina Hiseq-2500 platform to generate 125 bp pair-end reads (BIG, Shenzhen, CA, CHN). Totally, we obtained about 7 G high quality reads, which were aligned to chloroplast genomes of six Citrus species according to Wu et al. (Citation2019) and Xu et al. (Citation2019). Chloroplast genome assembly and annotation were performed according to the method described by Xu et al. (Citation2019), and the annotated chloroplast genome has been deposited in Genbank with the accession number MT880606.
The complete chloroplast genome of C. cavaleriei is 1,60,996 bp in size, containing a large single-copy region of 87,634 bp, a small single-copy region of 18,762 bp, and a pair of inverted repeat regions of 27,300 bp. Sequence annotation identified 114 unique genes from the C. cavaleriei chloroplast genome, including 80 protein-coding genes, 30 tRNA genes and 4 rRNA genes. Among the 114 unique genes, nine protein-coding genes (i.e. ndhB, rpl2, rpl22, rpl23, rps7, rps12, rps19, ycf2 and ycf15), seven tRNA genes (i.e. trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG and trnV-GAC) and all the 4 rRNA genes (rrn4.5, rrn5, rrn16 and rrn23 rRNAs) occur in double copies, and others occur as a single copy. The overall nucleotide composition of the chloroplast genome is: 30.47% A, 31.07% T, 19.58% C, and 18.88% G, with the total GC content of 38.46%.
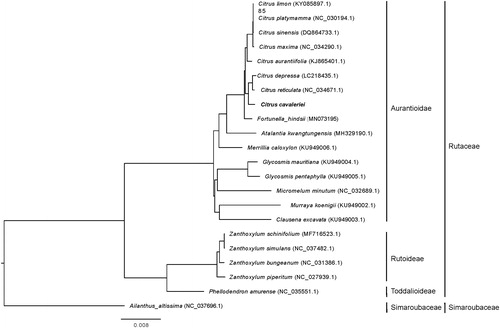
By using the complete chloroplast genomes of C. cavaleriei, 20 plant species from Rutaceae family and Ailanthus altissima (as outgroup), a maximum likelihood phylogenetic tree (Xu et al. Citation2019). Results showed that the relationship between the C. cavaleriei and C. reticulata chloroplasts is the closest (), which consistently support their chloroplast relationship that has been put forward by many scientists (Nicolosi et al. Citation2000; Penjor et al. Citation2013; Yang et al. Citation2017).
Figure 1. The maximum-likelihood phylogenetic tree constructed using the complete chloroplast genome sequences of Citrus cavaleriei and 20 plant species from the Rutaceae with Ailanthus altissima as outgroup. The phylogenetic tree was constructed according to the method and result described by Xu et al. (Citation2019).
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The raw data that support the findings of this study are openly available in NCBI Sequence Read Archive at [http://www.ncbi.nlm.nih.gov/bioproject/658640] under the BioProject ID PRJNA658640 and the annotated chloroplast genome has been deposited in Genbank [https://www.ncbi.nlm.nih.gov/genbank/] under the reference number MT880606.
Additional information
Funding
References
- Chen HM, Jiang D, Hu ZR, Li KM, He YR, Chen SC, Li LH, Zheng DS, Liu X. 2012. A newly discovered wild Ichang papeda population in Yuanjiang county, Yunnan Province. J Plant Genet Resour. 13:929–935. (in Chinese)
- Nicolosi E, Deng ZN, Gentile A, Malfa SL, Continella G, Tribulato E. 2000. Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor Appl Genet. 100:1155–1166.
- Penjor T, Yamamoto M, Uehara M, Ide M, Matsumoto N, Matsumoto R, Nagano Y. 2013. Phylogenetic relationships of citrus and its relatives based on matK gene sequences. PLOS One. 8(4):e62574.
- Wu GA, Terol J, Ibanez V, López-García A, Pérez-Román E, Borredá C, Domingo C, Tadeo FR, Carbonell-Caballero J, Alonso R, et al. 2018. Genomics of the origin and evolution of citrus. Nature. 554(7692):311–316.
- Wu JW, Liu F, Tian N, Liu JP, Shi XB, Bei XJ, Cheng CZ. 2019. Characterisation of the complete chloroplast genome of Fortunella Crassifolia Swingle and phylogenetic relationships. Mitochondr DNA B. 4(2):3538–3539.
- Xie X, Yang L, Liu F, Tian N, Che J, Jin S, Zhang Y, Cheng C. 2020. Establishment and optimization of Valencia sweet orange in planta transformation system. Acta Horticulturae Sinica. 47:111–119. (in Chinese)
- Xu S-R, Zhang Y-Y, Liu F, Tian N, Pan D-M, Bei X-J, Cheng C-Z. 2019. Characterization of the complete chloroplast genome of the Hongkong kumquat (Fortunella hindsii swingle). Mitochondr DNA B. 4(2):2612–2613.
- Yang X, Li H, Yu H, Chai L, Xu Q, Deng X. 2017. Molecular phylogeography and population evolution analysis of Citrus cavaleriei (Rutaceae). Tree Genet Genome. 13:29.
