Abstract
Chimonanthus praecox, a deciduous shrub tree, is endemic to China and widely cultivated in the world as a popular garden and ornamental plant. Here, we have reported its complete chloroplast genome with a length of 153,181 bp, containing a large single copy (LSC) region of 86,916 bp, a small single copy (SSC) region of 19,767 bp and two identical inverted repeat regions (IRs) of 23,249 bp. The overall GC contents of the plastome were 39.27%. A total of 114 unique genes were successfully annotated consisting of 80 protein-coding genes, 30 tRNA genes and four rRNA genes. Sixteen genes each possessed one intron and three genes had two introns. The ML phylogenetic analysis supports Chimonanthus as sister to Calycanthus. This result will be helpful for genetic breeding and population genetics of C. praecox, DNA barcoding of Chimonanthus, and phylogenetic studies of Calycanthaceae.
Chimonanthus praecox (linn.) Link, a deciduous shrub tree, is one of six species of the genus Chimonanthus native to China (Zhou et al. Citation2006). Now this species is widely cultivated in China and other temperate areas of the world as a popular garden and ornamental plant. Chimonanthus praecox is also used for medicine purposes, due to its extracts exhibiting multi-bioactivities (Ueyama et al. Citation1990; Zhang et al. Citation2009; Wang et al. Citation2011; Lou et al. Citation2018; Shu et al. Citation2019). In this study, we report and characterize the complete chloroplast genome of C. praecox might provide remarkable information for its molecular phylogeny and genetic breeding.
Fresh young leaves of C. praecox were collected from the same individual growing in the Institute of Botany, Jiangsu Province and Chinese Academy of Sciences (32.06˚N, 118.84˚E). A voucher specimen numbered FCM05 was deposited at the Herbarium of Institute of Botany, Jiangsu Province and Chinese Academy of Sciences. Genomic DNA was extracted from approximately 100 g fresh leaves followed the method of Ahmed and Fu (Citation2015). The DNA concentration and quality were assessed using a Qubit fluorometer (Invitrogen, San Diego, CA, USA) and a NanoDrop Spectrophotometer ND-1000 (NanoDrop Technologies, Wilmington, DE, USA), respectively, and then sequenced by Illumina Hiseq 2500 Sequencing platform (Illumina, Hayward, CA). Raw sequence reads have been deposited in the European Nucleotide Archive under BioProject ID PRJEB33250 (Accession no. ERS3550534). The generated raw data were de novo assembled using the program CLC Genomics Workbench v6.5 (CLC Bio, Aarhus, Denmark) with default parameters. The chloroplast genome was reconstructed by blasting all contigs against confamilial species Calycanthus floridus chloroplast genome (Accession no.: NC_004993) with the CLC BLAST tool. Annotations of the complete chloroplast genome of C. praecox were carried out using the online program GeSeq according to default values (Tillich et al. Citation2017), with Ca. floridus (Accession no.: NC_004993) as the reference, and adjusted manually in Geneious v6.0.3 (Biomatters Ltd, Auckland, New Zealand).
The annotated chloroplast genome was deposited at the GenBank database (Accession no. MT859152). The size of chloroplast genome was 153,181bp with the typical plastome structure of land plants, containing a large single copy (LSC) region of 86,916 bp, a small single copy (SSC) region of 19,767 bp and two identical inverted repeat regions (IRs: A and B) of 23,249 bp. However, the chloroplast genome of C. praecox was 71 bp smaller than that of Zhou’s submission (Accession no. NC_042744; Direct Deposit in GenBank). Additionally, 154 SNPs and 15 indels were found between the two C. praecox chloroplast genome sequences. Among the 154 SNPs, 58 SNPs were located in the exons of psaB (11), psaA (7), ycf2 (8), and ndhB (32); one SNP was located in the intron of trnS-CGA; 91 SNPs were located in the intergenic regions of ycf2-trnL-CAA (42), trnL-CAA-ndhB (34), trnA-UGC-rrna23 (14), and rps15-ycf1 (1); and four SNPs were located in trnL-CAA. There were 12 indels in introns, while only three indels in protein-coding genes (psaB and ycf2).
The overall GC content of the plastome is 39.27%, in which the corresponding values of the LSC, SSC, and IR region are 38.14%, 33.97%, and 43.65%, respectively. A total of 114 unique genes were successfully annotated consisting of 80 protein-coding genes, 30 tRNA genes and four rRNA genes. Among these genes, 16 genes (10 protein-coding and six tRNA genes) possessed one intron, while three genes (rps12, ycf3, clpP) had two introns. The rps12 gene was trans-spliced, with the 5′ exon located in the LSC region, and the 3′ exons duplicated in the IR region.
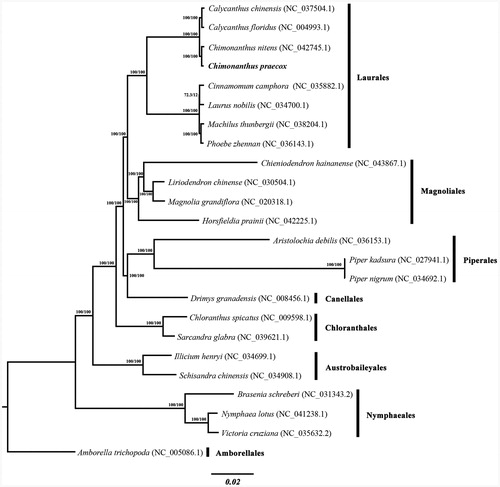
We used the complete chloroplast genome sequence of C. praecox and other 23 plastomes to construct a Maximum Likelihood tree through IQ-TREE based on the best-fit model estimated by ModelFinder (Nguyen et al. Citation2015; Kalyaanamoorthy et al. Citation2017); branch support values were assessed using UFBoot2 tests (Minh et al. Citation2013). The phylogenetic analysis indicated that C. praecox and C. nitens formed a clade of Chimonanthus, and the clade was subsequently sister to Calycanthus (), which is consistent with a previous study (Zhou et al. Citation2006). The new plastome sequence would provide a useful resource for genetic breeding and population genetics of this species, DNA barcoding of Chimonanthus, and the phylogenetic studies of Calycanthaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the European Nucleotide Archive under BioProject ID PRJEB33250 with accession No. ERS3550534 and GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, reference number MT859152.
Additional information
Funding
References
- Ahmed Z, Fu YB. 2015. An improved method with a wider applicability to isolate plant mitochondria for mtDNA extraction. Plant Methods. 11(1):56–56.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Lou H-Y, Zhang Y, Ma X-P, Jiang S, Wang X-P, Yi P, Liang G-Y, Wu H-M, Feng J, Jin F-Y, et al. 2018. Novel sesquiterpenoids isolated from Chimonanthus praecox and their antibacterial activities. Chin J Nat Med. 16(8):621–627.
- Minh BQ, Nguyen MAT, Von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Nguyen L, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Shu R, Wan Y, Wang X. 2019. Non-volatile constituents and pharmacology of Chimonanthus: a review. Chin J Nat Med. 17(3):161–186.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Ueyama Y, Hashimoto S, Nii H, Furukawa K. 1990. The volatile constituents of the flower concrete of Chimonanthus praecox Link. from China. Flavour Fragr J. 5(2):85–88.
- Wang W, Cao L, Xiong J, Xia G, Hu J. 2011. Constituents from Chimonanthus praecox (wintersweet). Phytochem Lett. 4(3):271–274.
- Zhang J-W, Gao J-M, Xu T, Zhang X-C, Ma Y-T, Jarussophon S, Konishi Y. 2009. Antifungal activity of alkaloids from the seeds of Chimonanthus praecox. Chem Biodivers. 6(6):838–845.
- Zhou S, Renner SS, Wen J. 2006. Molecular phylogeny and intra- and intercontinental biogeography of Calycanthaceae. Mol Phylogenet Evol. 39(1):1–15.