Abstract
Elaeidobius kamerunicus is the most important insect pollinator in oil palm plantations. In this study, the mitochondrial genome (mitogenome) of E. kamerunicus (17.729 bp), a member of the Curculionidae family, will be reported. The mitogenome consisted of 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), 2 ribosomal RNA genes (rRNAs), and a putative control region (CR). Phylogenetic analysis based on 13 protein-coding genes (PCGs) using maximum Likelihood (ML) methods indicated that E. kamerunicus belongs to the Curculionidae family. This mitochondrial genome provides essential information for understanding genetic populations, phylogenetics, molecular evolution, and other biological applications in this species.
Graphical Abstract
The oil palm pollinating weevil, Elaeidobius kamerunicus Faust. (Coleoptera: Curculionidae), is very important for the oil palm industry. Introduction of the weevil into oil palm plantations successfully improved fruit set and increased the yield of oil palm (Li et al. Citation2019). Recently, several genetic diversity studies of this species have been conducted using either nuclear or mitochondrial genetic information (Bakara et al. Citation2019, Citation2020; Haran et al. Citation2020; Tambunan et al. Citation2020). Nevertheless, the complete mitochondrial genome information of this species has never been reported before.
The present study provides the complete mitochondrial genome of the important oil palm pollinator E. kamerunicus, which is the first mitogenome sequenced for the tribe Derelomini. The samples were originally collected from an oil palm plantation of PT. Gunung Sejahtera Ibu Pertiwi, Kalimantan Tengah, Indonesia, with geospatial coordinate (2°25′28.6″S 111°47′26.8″E). The samples have been identified according to key morphological characteristics and deposited in the Insect specimen room of Bogor Agricultural University, Bogor, Indonesia, with a specimen voucher of EK-PL-2019. Total genomic DNA from one male individual was extracted using the gSYNC DNA Extraction Kit (Geneaid). The sequence of the complete mitochondrial genome was obtained through Illumina NextSeq 500 and assembled using NOVOPlasty v. 3.8.2 software (Dierckxsens et al. Citation2017).
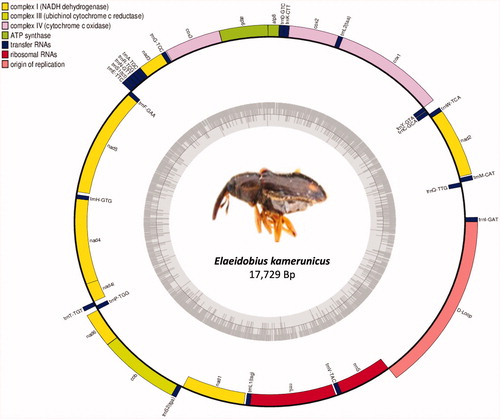
The complete mitogenome is a closed circular DNA molecule consisting of 13 PCGs, 22 tRNAs, 2 ribosomal RNA genes (rrnl and rrns), and 1 control region. The size of the complete mitochondrial genome was 17.729 bp. The sequence was deposited in the GenBank under accession number MT560591 or NC_049880. These two sequences are identical. Twenty-three genes (9 PCGs and 14 tRNA genes) are located on the majority strand (J-strand), with the remaining 14 genes (4 PCGs, 8 tRNAs, and 2 rRNAs) located on the minority strand (N-strand). Overall, the nucleotide composition of E. kamerunicus mitogenome consisted of 39.9% A, 33.6% T, 16.2% C, and 10.3% G with a highly biased A + T content of 73.5%. The whole mitogenome AT and GC skews are 0.09 and −0.22, respectively. The PCGs of the mitogenome encode a total of 3682 amino acids. All PCGs in this mitogenome start with common ATN codons. Stop codons in the PCGs included three types: TAA, TAG, or T––. The two ribosomal RNA genes (rrnl and rrns) encoding the large and small ribosomal subunits are located between trnL1 and trnV and between trnV and the control region, respectively. All of the 22 tRNAs ranged from 63 to 71 bp with a total length of 1.458 bp. Most of the tRNA genes were folded into a cloverleaf secondary structure, except for trnS, which lacks the DHU arm, which is commonly found in metazoan mitochondrial genomes (Cameron Citation2014). The control region of E. kamerunicus was located between trnI and rrns genes with a length of 5.516 bp and A + T contents 75.8%.
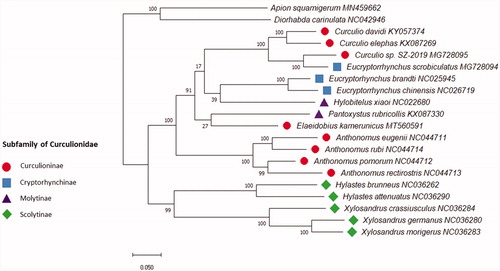
The results of phylogenetic analyses of E.kamerunicus based on sequences of 13 PCGs using Maximum Likelihood (ML) methods, support the placement of E.kamerunicus as a member of the Curculionidae family (). The mitogenome sequence of Apion squamigerum (Brentidae) and Diorhabda carinulata (Chrysomelidae) were used as outgroup. A recent study using phylogenomic data also showed that Curculioninae, Molytinae, and Cryptorhynchinae subfamilies are not monophyletic (Shin et al. Citation2018). Some of the classifications under family Curculionidae have been ambiguous, and subfamilies are still subject to change. In conclusion, the complete mitogenome of E. kamerunicus reported in this study provided important information for the phylogeny and evolution analysis of Curculionidae.
Acknowledgements
We thank all member of the Research and Development team of PT. Astra Agro Lestari Tbk, especially to Mr Adi Pancoro, Mr Satyoso Harjotedjo and Mr Cahyo Sri Wibowo. The author wishes to convey special thanks to Mr Santosa and Mr M. Hadi Sugeng, CEO and R&D Director of PT. Astra Agro Lestari Tbk, respectively.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that newly obtained at this study are available in the NCBI under accession number of MT560591 (https://www.ncbi.nlm.nih.gov/nuccore/MT560591) or NC_049880 (https://www.ncbi.nlm.nih.gov/nuccore/NC_049880).
Additional information
Funding
References
- Bakara RDJ, Buchori D, Kusumah RYM, Sahari B, Apriyanto A. 2019. Genetic diversity of Elaeidobius kamerunicus Faust. (Coleoptera: Curculionidae) across geographic areas in Indonesia: implications for oil palm sustainability. IOP Conf Ser Earth Environ Sci. 325:012003.
- Bakara RDJ, Tambunan VB, Apriyanto A, Kusumah RYM, Sahari B, Buchori D. 2020. Genetic diversity and population structure in Elaeidobius kamerunicus (Coleoptera: Curculionidae) inferred from mtDNA COI and microsatellite markers. Adv Biological Sci Res. 8:273–284.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Haran J, Ndzana Abanda RFX, Benoit L, Bakoumé C, Beaudoin-Ollivier L. 2020. Multilocus phylogeography of the world populations of Elaeidobius kamerunicus (Coleoptera, Curculionidae), pollinator of the palm Elaeis guineensis. Bull Entomol Res. 110(5):654–659.
- Li K, Tscharntke T, Saintes B, Buchori D, Grass I. 2019. Critical factors limiting pollination success in oil palm: a systematic review. Agric Ecosyst Environ. 280:152–160.
- Shin S, Clarke DJ, Lemmon AR, Moriarty Lemmon E, Aitken AL, Haddad S, Farrell BD, Marvaldi AE, Oberprieler RG, McKenna DD. 2018. Phylogenomic data yield new and robust insights into the phylogeny and evolution of weevils. Mol Biol Evol. 35(4):823–836.
- Tambunan VB, Apriyanto A, Ajambang W, Etta CE, Sahari B, Buchori D, Hidayat P. 2020. Molecular identification and population genetic study of Elaeidobius kamerunicus Faust. (Coleoptera: Curculionidae) from Indonesia, Malaysia and Cameroon based on mitochondrial gene. Biodiversitas. 21(7):3263–3270.