Abstract
Afla-Guard® is a commercial non-toxigenic Aspergillus flavus strain used to decrease aflatoxin contamination level in field. Its mitochondrial genome was sequenced, showing that its length is 29,208 bp with typical configuration of Aspergillus mitochondrial genome. 17 SNPs and 27 INDELs were identified by comparing with previous A. flavus mitochondrial genome. Phylogenetic trees present that A. flavus of Afla-Guard® was clustered with the previous A. flavus mitochondrial genome.
Afla-Guard® is a commercial product to decrease aflatoxin contamination level for improving quality of corns and peanuts (Wu et al. Citation2008; Durham et al. Citation2010; Dorner and Lamb Citation2006). This product contains Aspergillus flavus without aflatoxin gene cluster usually found in A. flavus (Abdel-Hadi et al. Citation2012). It can be used to understand phylogenetic position of nontoxic A. flavus. In addition, two mitochondrial genomes of A. flavus present that two genomes show different phylogenetic positions (data not shown), requiring additional mitochondrial genomes of A. flavus for clarifying this problem.
DNA of Afla-Guard® purchased from Syngenta was extracted using the optimized protocol (Lee et al. Citation2017). It was originally isolated from peanut seed at the USDA National Peanut Research Laboratory (Georgia, USA) in 1991 (NRRL 21882 in Agricultural Research Service Culture Collection). Raw data generated by HiSeq2500 and de novo assembly was conducted by Velvet 1.2.10 (Zerbino and Birney Citation2008). Gap filling was done by SOAPGapCloser 1.12 (Zhao et al. Citation2011) after confirming each base using BWA 0.7.17 and SAMtools 1.9 (Li et al. Citation2009; Li Citation2013). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used to annotate its mitogenome by comparing with those of A. flavus (JQ355000; Joardar et al. Citation2012).
The length of Afla-Guard® A. flavus mitogenome (GenBank accession is MT335777) is 29,208 bp, which is 3 bp longer than that of previously reported A. flavus (JQ355000). It is the 8th shortest mitogenome among 23 available Aspergillus mitogenomes (Park et al., in preparation). Numbers of PCGs, tRNAs, and rRNAs are 17, 27, and 2, respectively a typical configuration of Aspergillus mitogenomes.
Seventeen SNPs and 27 INDELs were identified as intraspecific variation on two A. flavus mitochondrial genomes. Two non-synonymous SNPs (nsSNPs) were identified in NAD1, two nsSNPs and three INDELs were found in hypothetical protein, and one nsSNP was in NAD4. Two synonymous SNPs (sSNPs) were found in COX1 and another two sSNPs were in NAD5. The remaining SNPs and INDELs were in intergenic space. Numbers of intraspecific variations are relatively large in comparison to those of Aspergillus oryzae, Aspergillus terrus, and Penicillium digitatum (Park et al., in preparation).
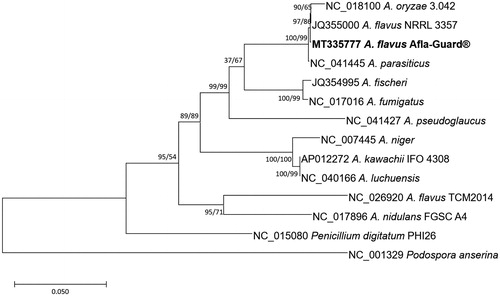
Sequence alignments of ten conserved genes from twelve Aspergillus and one Penicillium mitogenomes (Juhász et al. Citation2008; Futagami et al. Citation2011; Sun et al. Citation2011; Joardar et al. Citation2012; Zhao et al. Citation2012; Park, Kwon, Huang, et al. Citation2019; Park, Kwon, Zhu, Mageswari, Heo, Han, et al. Citation2019; Park, Kwon, Zhu, Mageswari, Heo, Kim, et al. Citation2019) including that of Afla-Guard® and one Podospora mitochondrial genome (Cummings et al. Citation1990) as an outgroup were calculated by MAFFT 7.450 (Katoh and Standley Citation2013) and concatenated. The neighbour-joining (10,000 bootstrap repeats) and maximum-likelihood (1,000 bootstrap repeats) phylogenetic trees were constructed using MEGA X (Kumar et al. Citation2018). Phylogenetic trees showed that two A. flavus mitochondrial genomes were clustered in one clade together with A. oryzae (). In addition, another A. flavus mitochondrial genome (NC_026920) was clustered with A. nidulans (), addressing its species identification.
Figure 1. Maximum-likelihood (bootstrap repeat is 1000) and neighbour-joining (bootstrap repeat is 10,000) phylogenetic trees of twelve Aspergillus, one Penicillium mitochondrial genome, and Podospora mitochondrial genome as an outgroup: Aspergillus flavus (MT335777 in this study, JQ355000, and NC_026920), Aspergillus oryzae (NC_018100), Aspergillus parasiticus (NC_041445), Aspergillus fischeri (JQ354995), Aspergillus fumigatus (NC_017016), Aspergillus pseudoglaucus (NC_041427), Aspergillus niger (NC_007445), Aspergillus kawachii (AP012272), Aspergillus luchuensis (NC_040166), Aspergillus nidulans (NC_017896), Penicillium digitatum (NC_015080), and Podospora anserina (NC_001329). Phylogenetic tree was drawn based on maximum-likelihood phylogenetic tree. The numbers above or below branches indicate bootstrap support values of maximum likihood and neighbour-joining phylogenetic trees, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Data availability statement
The sequence can be accessed via accession number MT335777 in NCBI GenBank. https://www.ncbi.nlm.nih.gov/genbank/.
Additional information
Funding
References
- Abdel-Hadi A, Schmidt-Heydt M, Parra R, Geisen R, Magan N. 2012. A systems approach to model the relationship between aflatoxin gene cluster expression, environmental factors, growth and toxin production by Aspergillus flavus. J R Soc Interface. 9(69):757–767.
- Cummings DJ, McNally KL, Domenico JM, Matsuura ET. 1990. The complete DNA sequence of the mitochondrial genome of Podospora anserina. Curr Genet. 17(5):375–402.
- Dorner JW, Lamb MC. 2006. Development and commercial use of afla-guard, an aflatoxin biocontrol agent. Mycotoxin Research. 22(1):33–38.
- Durham S, Suszkiw J, Flores A. 2010. Protecting corn crops from aflatoxin. Agric Res. 58(8):8.
- Futagami T, Mori K, Yamashita A, Wada S, Kajiwara Y, Takashita H, Omori T, Takegawa K, Tashiro K, Kuhara S. 2011. Genome sequence of the white koji mold Aspergillus kawachii IFO 4308, used for brewing the Japanese Distilled Spirit Shochu. Eukaryot Cell. 10(11):1586–1587.
- Joardar V, Abrams NF, Hostetler J, Paukstelis PJ, Pakala S, Pakala SB, Zafar N, Abolude OO, Payne G, Andrianopoulos A, et al. 2012. Sequencing of mitochondrial genomes of nine Aspergillus and Penicillium species identifies mobile introns and accessory genes as main sources of genome size variability. BMC Genomics. 13(1):698
- Juhász Á, Pfeiffer I, Keszthelyi A, Kucsera J, Vágvölgyi C, Hamari Z. 2008. Comparative analysis of the complete mitochondrial genomes of Aspergillus niger mtDNA type 1a and Aspergillus tubingensis mtDNA type 2b. FEMS Microbiol Lett. 281(1):51–57.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lee M-K, Park H-S, Han K-H, Hong S-B, Yu J-H. 2017. High molecular weight genomic DNA mini-prep for filamentous fungi. Fungal Genet Biol. 104:1–5.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN].
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Park J, Kwon W, Huang X, Mageswari A, Heo I-B, Han K-H, Hong S-B. 2019. Complete mitochondrial genome sequence of a xerophilic fungus, Aspergillus pseudoglaucus. Mitochondrial DNA B. 4(2):2422–2423.
- Park J, Kwon W, Zhu B, Mageswari A, Heo I-B, Han K-H, Hong S-B. 2019. Complete mitochondrial genome sequence of the food fermentation fungus, Aspergillus luchuensis. Mitochondrial DNA B. 4(1):945–946.
- Park J, Kwon W, Zhu B, Mageswari A, Heo I-B, Kim J-H, Han K-H, Hong S-B. 2019. Complete mitochondrial genome sequence of an aflatoxin B and G producing fungus, Aspergillus parasiticus. Mitochondrial DNA B. 4(1):947–948.
- Sun X, Li H, Yu D. 2011. Complete mitochondrial genome sequence of the phytopathogenic fungus Penicillium digitatum and comparative analysis of closely related species. FEMS Microbiol Lett. 323(1):29–34.
- Wu F, Liu Y, Bhatnagar D. 2008. Cost-effectiveness of aflatoxin control methods: economic incentives. Toxin Reviews. 27(3–4):203–225.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao G, Yao Y, Qi W, Wang C, Hou L, Zeng B, Cao X. 2012. Draft genome sequence of Aspergillus oryzae strain 3.042. Eukaryot Cell. 11(9):1178.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(Suppl 14):S2.
