Abstract
Plants in the genus Syzygium have been widely used as traditional medicine, fruit crops, and ornamental trees. In this study, we reported the complete chloroplast genome of Syzygium jambos (L.) Alston which was known as rose apple. The chloroplast genome of this species is 158541 bp in length, including a pair of inverted repeat regions (IRs) (26076 bp) that is divided by a large single copy area (LSC) (88036 bp) and a small single copy area (SSC) (18353 bp). The circular chloroplast genome of S. jambos contains 132 unique genes, composing of 85 protein-coding genes, 39 tRNA genes and 8 rRNA genes. Phylogenetic analysis indicates that S. jambos is clustered with species in genus Syzygium. This complete chloroplast genome of S. jambos will provide a powerful tool to accelerate breeding, biotechnological and phylogenetic study.
Syzygium Gaertn. is the largest genus in Myrtaceae, comprises about 1200 species and is distributed in the Old World tropics and subtropics (Biffin et al. Citation2010; Soh and Parnell Citation2015). Some species of genus Syzygium have been widely used as fruit crops (e.g. S. samarangense; Hao et al. Citation2016), ornamental trees (e.g. S. cumini; Abreu-Harbich et al. Citation2015) and traditional herbal medicines (e.g. S. aromaticum; El-Shouny et al. Citation2020). Syzygium jambos (L.) Alston, which is known as rose apple, is one of the most important species in Syzygium. It is native to southeast Asia and is cultivated in some areas of the tropics (Lim Citation2012). S. jambos has been reported as medicines to treat diabetes, inflammation and gastrointestinal disorders (Murugan et al. Citation2011; Rezende et al. Citation2013) for its leaf and fruit extracts containing high concentrations of tannins, phenolic acids, and other antioxidants (Oliveira et al. Citation2005; Gavillán-Suárez et al. Citation2015). Edible fruits of this species have a high percentage of pulp and contain an attractive aroma (roselike odour) and taste (sweet and slightly acidic), and are often used to make juices, jellies and jams (Guedes et al. Citation2004). In addition, S. jambos is also planted as ornamental trees in many countries for its outstanding adaptability. Most studies of S. jambos have focused on its chemical extraction and pharmacological properties. However, few genomic resources have been reported in this species, except a few gene sequences in phylogenetic analysis (Biffin et al. Citation2006).
Chloroplast genes and conserved sequences are often utilized for phylogenetic analysis and domestication studies of higher plants (Jansen et al. Citation2007). The whole chloroplast genome sequences have also been demonstrated the potential to understand structure and functional evolution (Jansen et al. Citation2007; Moore et al. Citation2010). In genus Syzygium, the chloroplast genome of some species such as S. samarangense and S. forrestii has been reported (Liu et al. Citation2018; Zhang et al. Citation2019), but the chloroplast genome of S. jambos has not been reported. Here, we sequenced and analyzed the complete chloroplast genome sequence of S. jambos based on the Illumina sequencing data. This study aimed to characterize the complete chloroplast genome sequence of S. jambos as a resource for future genetic studies.
Fresh leaves of S. jambos were collected from South China Botanical Garden, Chinese Academy of Sciences (Guangzhou, China). Total genomic DNA was extracted for library construction and sequencing. Voucher specimens of S. jambos were deposited at the herbarium of South China Botanical Garden (accession number: SCBG-CF-2061). The library was constructed with the insertion size of 350 bp. The high-throughput sequencing (pair-end 150 bp) was performed on an Illumina XTen platform. The clean reads were assembled by using the program NOVOPlasty (Dierckxsens et al. Citation2017). A ribulose-1, 5-bisphosphate carboxylase/oxygenase (rbcL) gene sequence from S. samarangense (GenBank accession: MH371141) was used as seed sequence, and the whole chloroplast genome sequences of S. samarangense and S. forrestii (MK102721) were used as a reference to resolve the inverted repeat in the chloroplast genome of S. jambos. The assembled chloroplast genome was annotated by the combination of PGA (Qu et al. Citation2019) and GeSeq (Tillich et al. Citation2017). For necessary genes, we manually corrected their positions of start and stop codons and boundaries between exons and introns. The annotated chloroplast genomic sequence has been deposited in GenBank with an accession number: MT731620.
The complete chloroplast genome of S. jambos is 158,541 bp in length, and has a typical quadripartite construction, which contains two inverted repeat regions (IRa and IRb) of 26,076 bp that is insulated by a large single-copy (LSC, 88,036 bp) and a small single-copy (SSC, 18,353 bp). The total GC content of complete chloroplast genome, LSC, SSC, IR regions is 37.0%, 55.5%, 11.6% and 32.9%, respectively. The complete chloroplast genome of S. jambos contains 132 unique genes, including 85 protein-coding genes, 39 tRNA genes and 8 rRNA genes. Introns are present in 18 of the annotated genes. Three of the intron containing genes (clpP, rps12, and ycf3) contain three exons. Most of these genes are single-copy genes. However, 18 genes were duplicated in IR regions.
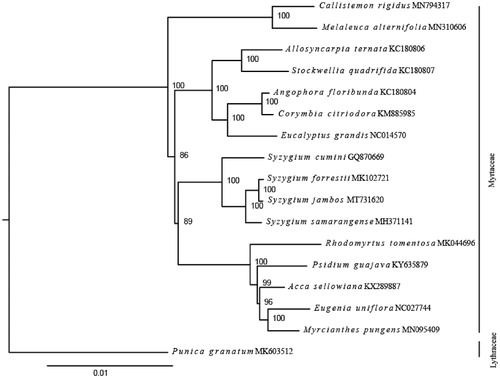
To confirm the phylogenetic position of S. jambos, the complete chloroplast genomes of 15 published species within Myrtaceae and one outgroup (Punica granatum, Lythraceae, MK603512) were downloaded from the NCBI GenBank database. Ninety-four chloroplast genes shared by all species we analyzed were extracted, and were aligned by using MUSCLE (Edgar Citation2004). We concatenated these genes and then constructed a maximum likelihood tree () using IQ-TREE (Nguyen et al. Citation2015). Phylogenetic analysis strongly supported that S. jambos was closely related to species in genus Syzygium (), which is consistent with the previous studies in Myrtaceae (Biffin et al. Citation2010; Thornhill et al. Citation2015). In conclusion, this published S. jambos chloroplast genome will provide a solid foundation for phylogenetic and evolutionary studies in Syzygium and is expected to improving the understanding of molecular mechanisms under pharmacological properties of S. jambos.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The raw sequencing data of S. jambos have been deposited in the NCBI Sequence Read Archive under accession numbers PRJNA658704. The chloroplast genome of the S. jambos was submitted to GenBank under accession number: MT731620. Treefile of 18 species and genes for phylogenetic analysis were deposited at Figshare: https://doi.org/10.6084/m9.figshare.12818804.v2.
Additional information
Funding
References
- Abreu-Harbich LV, Labaki LC, Matzarakis A. 2015. Effect of tree planting design and tree species on human thermal comfort in the tropics. Landscape Urban Plan. 138:99–109.
- Biffin E, Craven LA, Crisp MD, Gadek PA. 2006. Molecular systematics of Syzygium and allied genera (Myrtaceae): evidence from the chloroplast genome. Taxon. 55(1):79–94.
- Biffin E, Lucas E, Craven L, Ribeiro da Costa I, Harrington M, Crisp M. 2010. Evolution of exceptional species richness among lineages of fleshy-fruited Myrtaceae. Ann Bot. 106(1):79–93.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- El-Shouny WA, Ali SS, Hegazy HM, Elnabi MKA, Ali A, Sun J. 2020. Syzygium aromaticum L.: Traditional herbal medicine against cagA and vacA toxin genes-producing drug resistant Helicobacter pylori. J Tradit Complement Med. 10(4):366–377.
- Gavillán-Suárez J, Aguilar-Perez A, Rivera-Ortiz N, Rodríguez-Tirado K, Figueroa-Cuilan W, Morales-Santiago L, Maldonado-Martínez G, Cubano LA, Martínez-Montemayor MM. 2015. Chemical profile and in vivo hypoglycemic effects of Syzygium jambos, Costus speciosus and Tapeinochilos ananassae plant extracts used as diabetes adjuvants in Puerto Rico. BMC Complem Altern M. 15:244.
- Guedes CM, Pinto AB, Moreira RFA, Maria CAB. 2004. Study of the aroma compounds of rose apple (Syzygium jambos Alston) fruit from Brazil. Eur Food Res Technol. 219:460–464.
- Hao Y, Chen F, Wu G, Gao W. 2016. Impact of postharvest nitric oxide treatment on lignin biosynthesisrelated genes in wax apple (Syzygium samarangense) fruit. J Agric Food Chem. 64(45):8483–8490.
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci Usa. 104(49):19369–19374.
- Lim TK. 2012. Myrtaceae, Syzygium jambos. In Edible medicinal and non-medicinal plants: fruits. Vol. 3. London, New York: Dordrecht Heidelberg; p. 765.
- Liu J, Ni S, Zheng C, Shi C, Niu Y. 2018. Chloroplast genome of tropical and sub-tropical fruit tree Syzygium samarangense (Myrtaceae). Mitochondrial DNA B. 3(2):890–891.
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci Usa. 107(10):4623–4628.
- Murugan S, Uma Devi P, Parameswari NK, Mani KR. 2011. Antimicrobial activity of Syzygium jambos against selected human pathogens. Int J Pharma Pharmaceut Sci. 3:45–47.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Oliveira AC, Endringer DC, Amorim LA, das Gracas LBM, Coelho MM. 2005. Effect of the extracts and fractions of Baccharis trimera and Syzygium cumini on glycaemia of diabetic and non-diabetic mice. J Ethnopharmacol. 102(3):465–469.
- Qu X, Moore MJ, Li D, Yi T. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15: 50.
- Rezende WP, Borges LL, Alves NM, Ferri PH, Paula JR. 2013. Chemical variability in the essential oils from leaves of Syzygium jambos. Revista Brasileira de Farmacognosia. 23(3):433–440.
- Soh WK, Parnell J. 2015. A revision of Syzygium Gaertn. (Myrtaceae) in Indochina (Cambodia, Laos and Vietnam). Adansonia, Sér. 37:179–275.
- Thornhill AH, Ho SY, Kulheim C, Crisp MD. 2015. Interpreting the modern distribution of Myrtaceae using a dated molecular phylogeny. Mol Phylogenet Evol. 93:29–43.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zhang X, Wang J, Wang H, Zhao K, Zhu Z, Wang H. 2019. Complete plastome sequence of Syzygium forrestii Merr. et Perry (Myrtaceae): an endemic species in China. Mitochondrial DNA B. 4(1):126–127.