Abstract
Panisea was a small genus in Orchidaceae including 12 species, while its generic relationships had been unclear. Here, we report the complete chloroplast (cp) genome sequence and the features of Panisea uniflora to provide molecular data for further systematic research. This complete cp genome was 159,341 bp in length and presented a typical quadripartite structure including one large single-copy region (LSC, 87,166 bp), one small single-copy region (SSC, 18,781 bp), and two inverted repeat regions (IR, 26,697 bp, each). The cp genome encoded 132 genes, of which 113 were unique genes. Besides, a maximum-likelihood phylogenetic analysis indicated that P. uniflora was mostly related to Pholidota imbricata in tribe Arethuseae.
Panisea (Lindl.) Steud. was a small genus of Orchidaceae (Coelogyninae, Arethuseae, Epidendroideae) including about 12 species that were distributed from the Indian subcontinent to Southeast Asia (Subedi et al. Citation2011; Huang et al. Citation2012; Chase et al. Citation2015). There were five Panisea species in China, containing one endemic species (Chen and Jeffrey Citation2009). Since the generic relationships of Panisea within the subtribe Coelogyninae have been unclear (Gravendeel et al. Citation2001), the complete chloroplast (cp) genome of Panisea uniflora (Lindl.) Lindl. was reported for a better understanding of its phylogeny.
Leaf samples of P. uniflora were obtained from Fumin County, Yunnan Province, China (25°20′19′′N, 102°27′26′′E). The specimen was deposited in the Herbarium of Southwest Forestry University (HSFU, Lilu20180006). Total genomic DNA was extracted from fresh leaves using the modified CTAB procedure (Doyle and Doyle Citation1987) and sequenced on Illumina Hiseq 2500 platform (Illumina, San Diego, CA) at Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). We assembled the complete cp genome from the clean reads by the GetOrganelle pipe-line (Jin et al. Citation2018) and annotated the new sequences using the Geneious Prime version 2020.0.4 (Kearse et al. Citation2012). Finally, the complete cp genome sequence was submitted to GenBank with an accession number MT610371.
The complete cp genome sequence of P. uniflora was 1,59,341 bp in length and consisted of a large (LSC, 87,166 bp) and a small (SSC, 18,781 bp) single-copy regions, separated by a pair of identical inverted repeats (IR, 26,697 bp, each). The cp genome encoded 132 genes, of which 113 were unique genes (79 protein-coding genes, 30 tRNAs, and 4 rRNAs). The overall GC content was 37.3%, whereas the corresponding values of the LSC, SSC, and IR regions reaching 35.2, 30.1, and 43.3%, respectively.
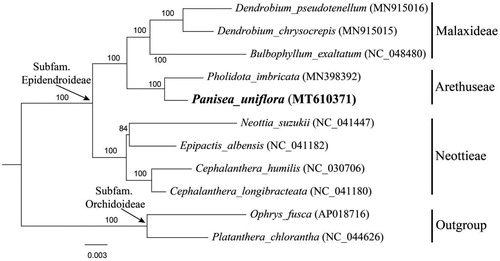
To confirm the phylogenetic position of P. uniflora, a maximum-likelihood (ML) tree was constructed based on 74 protein-coding genes with 11 complete cp genomes of related species. Eight species from three related tribes (Malaxideae Lindl., Arethuseae Lindl. and Neottieae Lindl.) in subfamily Epidendroideae were selected as ingroup, while two species from subfamily Orchidoideae as outgroup according to the updated molecular phylogeny of Orchidaceae (Chase et al. Citation2015; Li et al. Citation2016). All the sequences were downloaded from NCBI GenBank and aligned using MAFFT v7.307 (Katoh and Standley Citation2013). The maximum-likelihood (ML) analysis was performed using the CIPRES Science Gateway web server (RAxML-HPC2 on XSEDE 8.2.10) with 1000 bootstrap, replicates, and settings as described by Stamatakis et al. (Citation2008). It showed that P. uniflora was related to Pholidota imbricata Hook. with strong support (). This newly reported cp genome provides a good foundation for better understanding the tribal relationships within subfamily Epidenderoideae.
Acknowledgments
We are grateful to Dr. Fei Zhao in Kunming Institute of Botany Chinese Academy of Sciences, for his help in this manuscript. We also thank Mr. Zhi-Feng Xu and Mrs. Xiao-Yun Wang in the Wild Orchid Conservation Center of Yunnan Fengchunfang Biotechnology Company, for providing sample materials.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT610371.
Additional information
Funding
References
- Chase MW, Cameron KM, Freudenstein JV, Pridgeon AM, Salazar G, Berg CVD, Schuiteman A. 2015. An updated classification of Orchidaceae. Bot J Linn Soc. 177(2):151–174.
- Chen SC, Jeffrey JW. 2009. Panisea (Lindley) Lindley. In: Wu ZY, Raven PH, Hong D, editors. Flora of China. 25. Beijing (China): Science Press; St. Louis (MO): Botanical Garden Press. p. 333–334.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gravendeel B, Chase MW, Vogel EFD, Roos MC, Mes THM, Bachmann K. 2001. Molecular phylogeny of Coelogyne (Epidendroideae; Orchidaceae) based on plastid RFLPS, matK, and nuclear ribosomal ITS sequences: evidence for polyphyly. Am J Bot. 88(10):1915–1927.
- Huang MZ, Yin JM, Yang GS, Tan YH. 2012. Panisea moi, a new species (Orchidaceae: Epidendroideae) from Hainan, China. Phytotaxa. 60(1):13–16.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li MH, Zhang GQ, Lan SR, Liu ZJ. 2016. A molecular phylogeny of Chinese orchids. J Sytematics Evol. 54(4):349–362.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web-servers. Syst Biol. 75:758–771.
- Subedi A, Chaudhary RP, Vermeulen JJ, Gravendeel B. 2011. Panisea panchaseensis sp. nov. (Orchidaceae) from central Nepal. Nord J Bot. 29(3):361–365.