Abstract
We reported the first complete plastid genome of Aspidopterys (Malpighiaceae) in this study. The complete plastome of Aspidopterys obcordata is 160,453 bp in length with a base composition of A (31.4%), G (18.5%), C (18.2%), and T (32.0%). Structurally, the genome contains two short inverted repeats (26,905 bp for each), which are separated by a large single copy region (88,491 bp) and a small single copy region (18,152 bp). The plastome contained 113 unique genes, including 79 protein-coding genes, 30 transfer RNAs, and 4 ribosomal RNAs. Phylogenetic analyses showed that A. obcordata was sister to Bunchosia argentea in the monophyletic Malpighiaceae. This study provided a high-quality plastome sequence for future studies in Aspidopterys, as well as Malpighiaceae.
Aspidopterys obcordata Hemsl. (Malpighiaceae) is a woody liana and distributes in Hainan and South Yunnan, China (Chen and Funston Citation2008). The whole plant contained some important biochemical compounds having effects on antiphlogistic and diuresis treatments, which could be used to cure urinary infection and calculus, cystitis, rheumatism, and bone pain, as well as postpartum weak health and children’s digestion diseases (e.g., Wu et al. Citation2001; Li et al. Citation2016; Hu et al. Citation2018; Li et al. Citation2019). Plants of this species are important ingredients in some ancient prescriptions of the traditional Dai medicine (State Administration of Traditional Chinese Medicine Citation2005). To date, there had four complete chloroplast genomes reported representing three genera in Malpighiaceae, but none of them belongs to Aspidopterys A. Juss. ex Endl. In this study, we reported the complete plastome sequence of A. obcordata in the first time, which can be used to develop DNA markers for molecular authentication and conservation genetics of Aspidopterys.
Fresh leaves of A. obcordata (W.-B. Yu et al. DAI-159 (KUN)) were collected at Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences (21°55′18.6.6″N 101°16′32.09″E), Mengla, China, then was dried using silica-gel. Genomic DNA was extracted using a modified CTAB method (Doyle and Doyle Citation1987). The 150 bp pair-end reads were generated by Illumina Hi-Seq 2500 using 500 bp insert size library. Around 7.25 Gb clean data with 20,455,554 reads were de novo assembled using GetOrganelle toolkit (Jin et al. Citation2020). The plastome was annotated using GeSeq (Tillich et al. Citation2017), then manually adjusted in Geneious (Kearse et al. Citation2012).
The whole plastome of A. obcordata was 1,60,453 bp (MT590775) in length with two inverted repeats (IRs, 26,905 bp for each) that were separated by a large single-copy (LSC, 88,491 bp) and a small single-copy (SSC 18,152 bp) regions. The plastome contained 131 genes in total, including 113 unique genes in 79 protein-coding, 30 tRNA, and four rRNA genes. There are nine protein-coding (three are partial), seven tRNA, and four rRNA genes in IR regions. The overall GC content was 36.6%.
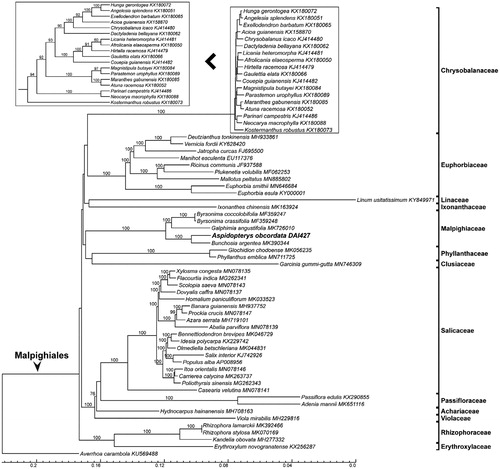
The whole sequences with one IR region of 62 taxa of Malpighiales and an outgroup Averrhoa carambola L. (Oxalidales, KU569488) were aligned using MAFFT (Katoh and Standley Citation2013), then the gaps were trimmed by trimAl (Capella-Gutiérrez et al. Citation2009) using the command ‘-gt 0.9 -cons 60.’ For Maximum Likelihood analyses, we used RAxML (Stamatakis et al. Citation2008) using GTRGAMMAI model with 1000 bootstraps to reconstruct phylogeny of Malpighiales. Phylogenetic analysis showed that the backbone of Malpighiales was not resolved, and A. obcordata was sister to Bunchosia argentea (Jacq.) DC. (MK390344) in the monophyletic Malpighiaceae (). This new plastome sequence will be valuable for investigations on systematics and conservation genetics of Aspidopterys.
Disclosure statement
The authors report no conflicts of interest and responsible for the content and writing of this article.
Data availability statement
The complete plastome was deposited at GenBank with accession number MT590775, which was also available at Figshare (doi: 10.6084/m9.figshare.12497900). The data was collected without violation of the protection of human subjects, or other valid ethical, privacy, or security concerns.
Additional information
Funding
References
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Chen H-K, Funston M. 2008. Malpighiaceae. In: Wu CY, Raven P, Hong DY, editors. Flora of China. Beijing & St. Louis: Science Press & Missouri Botanical Garden Press; p. 132–138.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry. 19:11–15.
- Hu M, Li Y, Sun Z, Huo X, Zhu N, Sun Z, Liu Y, Wu H, Xu X, Ma G, et al. 2018. New polyoxypregnane glycosides from Aspidopterys obcordata vines with antitumor activity. Fitoterapia. 129:203–209.
- Jin J-J, Yu W-B, Yang J-B, Song Y, Depamphilis C W, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21:241 doi:10.1186/s13059-020-02154-5. PMC: 32912315
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li Y, Li G, Song M, Li X, Zhan X, Lu J, Chen X. 2016. [Acute toxicity study of Aspidopterys obcordata aqueous extract in Sprague-Dawley rats]. J Tradit Chin Med. 36(3):377–381.
- Li Y, Ma G, Lv Y, Su J, Li G, Chen X. 2019. Efficacy of obcordata A from Aspidopterys obcordata on kidney stones by inhibiting NOX4 expression. Molecules (Basel, Switzerland). 24(10):1957.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57(5):758–771.
- State Administration of Traditional Chinese Medicine 2005. Chinese Materia Medica: Dai medicinal plants. Shanghai: Shanghai Scientific & Technical Publishers.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wu R, Ye Q, Chen N, Zhang G. 2001. Study on the chemical constituents of Aspidopterys obcordata Hemsl. Nat Prod Res Dev. 13:14–16.