Abstract
Camellia pingguoensis D. Fang is a shrub which is found on limestone of karst forests in Guangxi, China. In this study, we characterized the whole plastid genome of C. pingguoensis using Illumina paired-end sequencing reads. The plastome is 156,621 bp in length, containing two copies of inverted repeat (IR) regions (26,046 bp), a large-single copy (LSC) region (86,289 bp), and a small-single copy (SSC) region (18,240 bp). A total of 114 unique genes in the genome has 80 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The phylogenetic result indicates C. pingguoensis is closely related to C. nitidissima C. W. Chi.
Camellia L., comprising more than 200 species, is the type and the largest and economically most important genus in the family Theaceae (Vijayan et al. Citation2009; Huang et al. Citation2014). Most species of Camellia are distributed in southeastern and eastern Asia, China is the center of species diversity, possess more than 80% of the species (Gao et al. Citation2005). About 82% the species of the genus inhabit the subtropical (the subtropical evergreen broadleaved forests) EBLFs of East Asia (Yu et al. Citation2017). Camellia pingguoensis D. Fang is a shrub which grows between 1-3 meters tall with pale yellow flowers which fade to white when fully opened. It is found on limestone of karst forests between 100–500 meters above sea level in Guangxi (Wheeler Citation2015). This species is a valuable ornamental plant, and is classified as Endangered in the 2004 China Red List due to continuing population decline and limited distribution (Wheeler Citation2015). In this study, we characterized the whole plastid genome of C. pingguoensis.
Fresh leaves of an individual of C. pingguoensis was collected from South China Botanical Garden (23°10′58″N, 113°21′28″E), and used to extract total genome DNA using a modified CTAB protocol (Doyle and Doyle Citation1987). The voucher specimen was deposited at South China Botanical Garden Herbarium (voucher number: Wu19017). The Illumina paired-end library was prepared and sequenced based on Illumina Hiseq X Ten platform at Beijing Genomics Institute (Wuhan, China). The plastome was assembled using NOVOPlasty (Dierckxsens et al. Citation2017) with a reference plastome Camellia granthamiana Sealy (NC_038181). The draft plastid genome was checked by remapping reads in Geneious Prime 2019 (Biomatters, Ltd, Auckland, New Zealand). The coding genes were annotated and manually adjusted using Geneious Prime 2019, and tRNA were annotated using ARAGORN (Laslett and Canback Citation2004). Phylogenomic analysis was performed using protein-coding genes from 28 Camellia taxa and four outgroups in Theaceae. The aligned matrix was implemented in MAFFT (Katoh and Standley Citation2013). The maximum likelihood tree was constructed using RAxML (Stamatakis Citation2014) with 1000 rapid bootstrap replicates ().
Figure 1. Maximum likelihood tree of Camellia based on 79 common protein-coding genes. The node labels indicate the ML bootstrap (1000 replicates) support values.
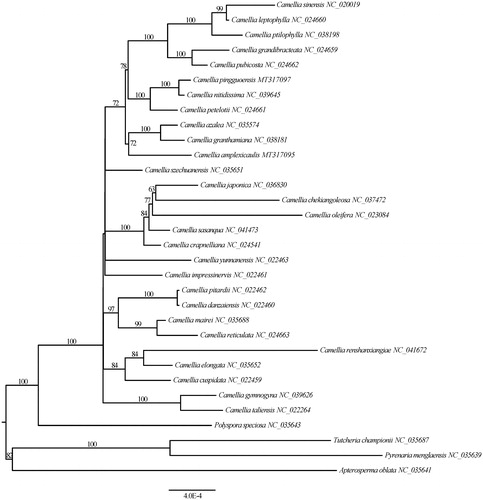
The plastome of C. pingguoensis (GenBank accession number MT317097) is 156,621 bp in length, consisting of two copies of inverted repeat (IR) regions (26,046 bp), a large-single copy (LSC) region (86,289 bp), and a small-single copy (SSC) region (18,240 bp). The plastome has a total of 114 unique genes, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. The GC values of the LSC, SSC, and IR regions are 35.4, 30.6, and 43%, respectively. The overall GC content of the plastome is 37.3%. The ML tree showed C. pingguoensis and Camellia nitidissima C. W. Chi were recovered as sister relationship with strong bootstrap support (100%).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/, reference number MT317097.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gao J, Parks CR., Du Y 2005. Collected species of the genus Camellia and illustrated outline. Zhejiang (China): Zhejiang Science and Technology Press.
- Huang H, Shi C, Liu Y, Mao SY, Gao LZ. 2014. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol. 14:151.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32:11–16.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Vijayan K, Zhang WJ, Tsou CH. 2009. Molecular taxonomy of Camellia (Theaceae) inferred from nrITS sequences. Am J Bot. 96:1348–1360.
- Wheeler L. 2015. Camellia pingguoensis. The IUCN Red List of Threatened Species 2015: e.T62056505A62056514. http://dx.doi.org/10.2305/IUCN.UK.2015-2.RLTS.T62056505A62056514.en
- Yu XQ, Gao LM, Soltis DE, Soltis PS, Yang JB, Fang L, Yang SX, Li DZ. 2017. Insights into the historical assembly of East Asian subtropical evergreen broadleaved forests revealed by the temporal history of the tea family. New Phytol. 215:1235–1248.
