Abstract
Hydrangea davidii, a perennial shrub of Hydrangeaceae, is an ornamental plant endemic to China. Here, we report the complete chloroplast genome of H. davidii. The complete chloroplast genome is totally 158,054 bp in length with a typical quadripartite structure. It consists a pair of inverted regions (IRs) of 26,140 bp, which were separated by a large single copy (LSC) region of 87,008 bp and a small single copy (SSC) region of 18,766 bp, respectively. The chloroplast genome encoded 131 genes, including 85 protein-coding genes, 8 rRNA genes and 38 tRNA genes. The GC content in the whole cp genome, LSC region, SSC region, and IR region are 37.8%, 36.0%, 31.7%, and 43.1%, respectively. In total, 49 SSRs were identified in the complete chloroplast genome. Phylogenetic analysis showed that H. davidii is closely related to Hydrangea platyarguta with a support rate of 100%.
The genus Hydrangea Linnaeus (Hydrangeaceae), including ca. 73 species, is mainly distributed in the temperate zone of the northern hemisphere with 33 species (25 endemic) in China (Wei and Bartholomew Citation2001). Hydrangea has been cultivated for gardening plants as well as pot-cultured and cut-flowers worldwide due to its high ornamental value. There are many unique and interesting characters of Hydrangea made it famous, such as big inflorescences with both inconspicuous fertile and attractive neuter flowers, flower color change with soil pH degree, long flowering period from June to August (Cerbah et al. Citation2001; Barbara Conolly et al. Citation2010; Yue and Peng Citation2019). H. davidii Franchet is a 1–3 m tall perennial shrub distributing in mixed forests on mountain slopes or in valleys of Sichuan, Yunnan and Guizhou of China (Wei and Bartholomew Citation2001). It has big and dense corymbose cymes with sterile flower borne at margin of inflorescence and has been cultivated for garden plants with high ornamental value. Additionally, it had been reported that H. davidii, as one of three origin plants for Traditional Chines Medicine Xiao-Tongcao, had medicinal value on anti-inflammatory and antipyretic and diuretic effects (Shen et al. Citation1998). In this study, we first report the complete chloroplast (cp) genome of H. davidii to provide genomic resource for further research on the phylogenetic reconstruction, genetic diversity and evolution.
The fresh leaves were collected from a healthy H. davidii individual, growing in Xiao-Yanfang Natural Reserve, Yongshan County (28°22'15″N, 110°55'22″E) in Yunnan Province. The voucher specimens (XYF-092) was deposited in the Herbarium of Yunnan Normal University. The total genomic DNA was extracted using a modified CTAB protocol (Allen et al. Citation2006). A pair-end (PE) sequence library was constructed and sequenced using the Illumina HiSeq 2500-PE150 platform (Illumina, USA). Raw sequence data were filtered to obtain clean reads using NGS QC Toolkit version 2.3.3 with default parameters (Patel and Jain Citation2012). The complete cp genome was de novo assembled by NOVOPlasty (Dierckxsens et al. Citation2016) and annotated with the online annotation tool GeSeq (Tillich et al. Citation2017). The SSRs were detected using online software MISA (Thiel et al. Citation2003). The unit sizes of mono-, di-, tri-, tetra-, penta-, and hexa-nucleotide repeats were set to minimum number of repeats of 10, 5, 4, 3, 3, and 3, respectively.
The complete chloroplast genome of H. davidii (GenBank accession number: MT861130) is 158,054 bp in length and has the typical quadripartite structure, including a large single copy (LSC) region of 87,008 bp, a small single copy (SSC) region of 18,766 bp, and two separated inverted region (IRs) of 26,140 bp. The overall GC content of the cp genome is 37.8%, while the corresponding values of the LSC, SSC, and IR regions were 36.0%, 31.7%, and 43.1%, respectively. The cp genome encoded 131 genes, including 85 protein-coding genes (PCGs), 8 rRNA genes and 38 tRNA genes. Among these genes, 61 PCGs and 23 tRNA genes are located in the LSC region (including one interregional gene rps19), while 12 PCGs and one tRNA gene occur in the SSC region (including two interregional genes ycf1 and ndhF). All the eight rRNA genes are duplicated in the IR regions. Each of the IR regions contains six PCGs and seven tRNA genes. A total of 16 genes contains a single intron, and two genes (ycf3 and clpP) contain two introns. 49 SSRs were identified in the complete chloroplast genome in total. The numbers of mono-, di-, tri-, tetra-, penta-, and hexa-nucleotide nucleotides SSRs are 40, 2, 2, 3, and 2, 0, respectively and mono-nucleotide A/T SSR motif was most frequent.
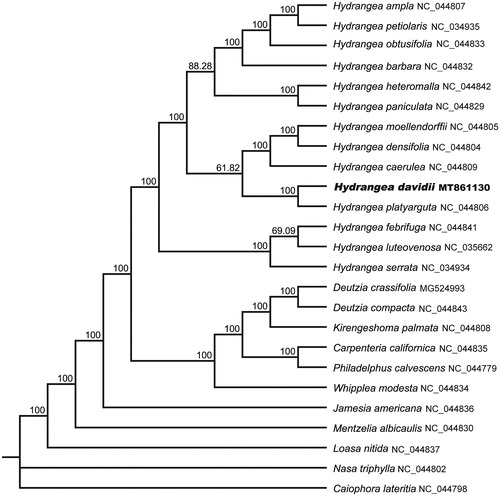
To analyze the phylogenetic location of H. davidii, 21 species of Hydrangeaceae and four outgroups were aligned by the MAFFT version 7 software (Katoh and Standley Citation2013). Maximum-likelihood (ML) tree was constructed using RAxML 8.2.11 (Stamatakis Citation2014) with the GTR + G nucleotide substitution model and all branch nodes were calculated under 1,000 bootstrap replicates. The phylogenetic analysis revealed that all sampled species of Hydrangea were clustered into one monophyletic clade with a high bootstrap value. Within Hydrangea, H. davidii is closely related to H. platyarguta with a support rate of 100% ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number [MT861130] [SRR12526392], or available from the corresponding author.
Additional information
Funding
References
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1(5):2320–2325.
- Barbara Conolly N, Bassuk NL, MacRae PF. 2010. Response of five Hydrangea species to foliar salt spray. J Environ Hort. 28(3):125–128.
- Cerbah M, Mortreau E, Brown S, Siljakyakovlev S, Bertrand H, Lambert C. 2001. Genome size variation and species relationships in the genus Hydrangea. Theor Appl Genet. 103(1):45–51.
- Dierckxsens N, Mardulyn P, Smits G. 2016. Novoplasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4).:e18.
- Katoh K, Standley DM. 2013. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Patel RK, Jain MK. 2012. NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619.
- Shen YJ, Zeng N, Jia M, Zhang Y, Wei TF, Ma YY. 1998. Experimental studies on anti-inflammatory, antipyretic and diuretic effects of several species of Tongcao and Xiao-Tongcao. Zhongguo Zhong Yao Za Zhi. 23(11):687.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Thiel T, Michalek W, Varshney RK, Graner A. 2003. Exploiting EST databases for the development and characterization of gene-derived ssr-markers in barley (Hordeum vulgare L.). Theor Appl Genet. 106(3):411–422.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. Geseq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wei ZF, Bartholomew B. 2001. Hydrangea. In: Wu CY, Raven PH, editors. Flora of China. Beijing and St Louis: Science Press and Missouri Botanical Garden Press; p. 411–422.
- Yue Y, Peng ZH. 2019. Construction of the thematic plant landscape with hydrangea in China. Architect Cul. 180(03):151–153.