Abstract
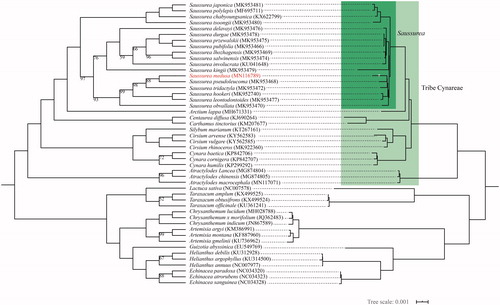
Saussurea medusa Maxim. is a subnival plant of Asteraceae with abundant medicinal and ecological value. Until now, few studies have been conducted on S. medusa, especially in phylogenetic relationships and species identification. The S. medusa cp genome had a typical quadripartite structure with a conserved genome arrangement. It was 152,500 bp in size, consisting of a large single copy (LSC) region of 83,553 bp and a small single copy (SSC) region of 18,545 bp, separated by a pair of inverted repeats (IRs) of 25,201 bp. It contained 113 unique genes, including 80 protein-coding genes (PCGs), 29 tRNA and four rRNA genes. The overall GC content was 37.67%. Moreover, a phylogenetic analysis, based on 48 complete cp genomes using Maximum Likelihood (ML) method, indicated that S. medusa was relatively closed to S. pseudoleucoma and was well-clustered within genus Saussurea.
Keywords:
S. medusa is a rare subnival plant known as “snow lotus” that belongs to the genus Saussurea of the Cynareae (Asteraceae) (based on APG IV system) (Byng et al. Citation2016). The genus Saussurea is a taxonomically complex and controversial group comprising approximately 460 species, of which 300 are present in China (Chen Citation2015). S. medusa is a monocarpic herb that occur on gravel slopes and alpine scree, typically at elevations of 3000–5600 m. The habitat of it is quite fragile and the population is fragmented and isolated. Owing to its ability of tolerating multiple extreme stresses, such as strong ultraviolet radiation, cold, hypoxia and drought, this herb is expected to provide new insights for the study of alpine plants.
In the present study, the fresh leaves of S. medusa were collected from Reshuidaban, Qinghai Province, China. A voucher specimen (QPMB 0334412) of S. medusa has been deposited in the Qinghai-Tibetan Plateau Museum of Biology (QPMB), Northwest Institute of Plateau Biology, Chinese Academy of Sciences (CAS). Total genomic DNA was extracted by modified CTAB method (Idress Hamad Citation2011). Then the cp genome was sequenced using Illumina HiSeq platform provided by Genepioneer Biotechnologies (Nanjing 210014, China). Approximately, 10,171,459,200 clean reads and 33,904,864 pair-end reads were obtained, with a quality value ≥ Q30, accounting for 92.19%. The SPAdes (Bankevich et al. Citation2012) was used to genome assembly, based on the remaining clean data. The annotation of rRNAs, tRNAs, and protein-coding genes of the cp was performed using the HMMER (Wheeler & Eddy Citation2013), ARAGORN (Laslett & Canback Citation2004) and BLAST (Boratyn et al. Citation2013; McGinnis & Madden Citation2004), respectively. Phylogenetic trees were constructed by Maximum Likelihood (ML) methods using the IQ-tree version 1.6.12 (Trifinopoulos et al. Citation2016) with the best model of TVM + F + R6, of which the bootstrap values of 1000 replicates to assess the branch support. Then the tree was visualized and optimized by iTOL online tool (Letunic & Bork Citation2016).
The complete cp genome of S. medusa with gene annotation was deposited in NCBI GenBank with accession number MN116789. The complete cp genome was 1,52,500 bp in length, and possessed the typical quadripartite structure of angiosperm, including a large single-copy (LSC) region of 83,553 bp and a small single-copy (SSC) region of 18,545 bp separated by a pair of inverted repeat (IR) regions of 25,201 bp. The overall GC content of the S. medusa cp genome was 37. 67%. Among the four regions, the GC content of the IRs (43.11%) was higher than it in LSC (35.78%) and SSC (31.42%). In this genome, 113 unique genes were identified, which contained 80 PCGs, 29 tRNA and four rRNA genes. There were 21 tRNA genes and 61 PCGs in LSC region, also, one tRNA gene and 12 PCGs in SSC region.
As a result of the phylogenetic analysis, S. medusa was well-positioned within the genus Saussurea and clustered together with S. pseudoleucoma (). Overall, our results in this study will contribute to further researches on phylogeny, radiation evolution of genus Saussurea.
Disclosure statement
The authors declare no conflict of interest.
Data availability statement
The raw data of sequencing is openly available in Figshare at https://figshare.com/articles/SRR9681919/12470378/1.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, et al. 2013. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41:W29–W33.
- Byng JW, Chase MW, Christenhusz MJM, Fay MF, Judd WS, Mabberley DJ, Sennikov AN, Soltis DE, Soltis PS, Stevens PF, et al. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Chen Y-S. 2015. Flora of Pan-Himalayas. Asteraceae II Saussurea. Cambridge: Cambridge University Press.
- Idress Hamad A. 2011. Modified CTAB method for high quality genomic DNA extraction from medicinal plants. Pak J Biol Sci. 21:998–999.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–245.
- McGinnis S, Madden TL. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32:W20–W25.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235.
- Wheeler TJ, Eddy SR. 2013. nhmmer: DNA homology search with profile HMMs. Bioinformatics. 29(19):2487–2489.