Abstract
Homatula berezowskii (Günther 1896), used to be recognized as a synonym of Homatula variegate, was now identified as a valid species. However, these morphological studies lack genetic evidence to support. In the present study, we determined the first complete mitochondrial genome of H. berezowskii by Sanger dideoxy sequencing. The genome size is 16,570 bp and it contains 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a non-coding control region (D-loop). Phylogenetic analyses based on the complete mitochondrial genome indicated the H. berezowskii was clustered with H. potanini first and then with the H. variegate. This work may be helpful to clarify the taxonomic status of H. berezowskii.
Homatula berezowskii (Günther 1896), an endemic freshwater fish to China, is only distributed in the upstream of the Yellow River and the Yangtze River, and their tributaries (Ding Citation1994). This fish used to be considered as a synonym of H. variegatewas recently identified as a valid species (Hu and Zhang Citation2010; Gu and Zhang Citation2012). However, most of the taxonomic studies of this fish were based on the morphological parameters, only a partial sequence of cytochrome b (cytb) was used to revise the classification of Homatula and Schistura in the Sichuan province of China (Xiong et al. Citation2017). To further clarify the taxonomic status of H. berezowskii, we sequenced its complete mitochondrial genome.
The Homatula berezowskii sample of H. berezowskii was collected from Qujing City (103.7°E, 25.6°N), Yunnan Province, China, and now is stored in Key Laboratory of Molecular Biology for Agriculture, Northwest A&F University, Yangling, China (voucher no. FS-2014-Y03). Total DNA was extracted by using the Animal Genomic DNA extraction Kit (Tiangen Biotech, Beijing, China). The whole mitochondrial genome was PCR amplified, Sanger-sequenced, and assembled following the method described by Zhou et al. (Citation2014).
The complete H. berezowskii mitochondrial genome (GenBank accession no. NC_040302.1) was 16,570 bp in length, including 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a non-coding control region (D-loop region). All the PCGs employ ATG as the start codon, except for COX1, using the start codon GTG. As for the termination codon, six genes (ND1, ND2, COX1, ND3, ND4, ND6) use TAG, and four genes (ATP8, ATP6, ND4L, ND5) use TAA as their stop codon. The remaining three PCGs use the incomplete stop codon T–. No TA– stop codon, which differs from Homatula potanini (Que et al. Citation2012) and Homatula variegatus (Shi et al. Citation2014).
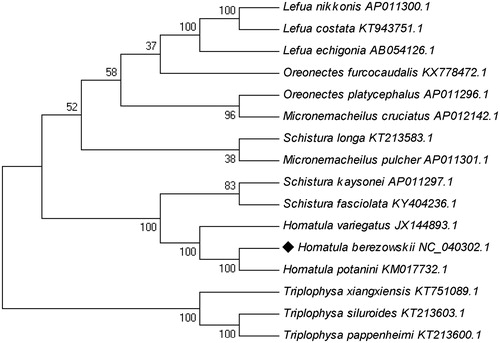
The phylogenetic tree was constructed with 16 fish species of the Nemachilinae subfamily by using the maximum likelihood (ML) method with 1000 bootstrap replicates. Results showed that H. berezowskiiis sister to H. potanini, and shares a recent common ancestor with H. variegatus (). The genetic data agree with the morphological data (Xiong et al. Citation2017). The Schistura spp., Triplophysa spp., and Lefua spp. were also clustered together, but Micronemacheilus spp. and Oreonectes spp. were intermingled with other species, suggesting further taxonomic investigations are needed.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number NC_040302.1.
Additional information
Funding
References
- Ding RH. 1994. The fishes of Sichuan. Chengdu: Sichuan Publishing House of Science and Technology.
- Gu JH, Zhang E. 2012. Homatula laxiclathra (Teleostei: Balitoridae), a new species of nemacheiline loach from the Yellow River drainage in Shaanxi Province, northern China. Environ Biol Fish. 94(4):591–599.
- Hu YT, Zhang E. 2010. Homatula pycnolepis, a new species of nemacheiline loach from the upper Mekong drainage, South China (Teleostei: Balitoridae). Ichthyol Explor Freshw. 21:51–62.
- Que YF, Tang HY, Xu N, Xu DM, Shi F, Yang Z, Li WT. 2012. Characterization of the complete mitochondrial genome sequence of Homatula potanini (Cypriniformes, Nemacheilidae, Nemachilinae). Mitochondr DNA. 27:1–3.
- Shi Y, Xing L, Chen T, YangM, You P. 2014. Sequencing and analysis of the complete mitochondrial genome of Paracobitis variegatus. JShaanxi Normal Univ (Nat Sci Ed). 42:50–56.
- Xiong K, Sun Z, Yanshu G. 2017. Revision of Homatula and Schistura classification in Sichuan Province. J China West Normal Univ (Nat Sci). 38:21–27.
- Zhou X, Yu Y, Li Y, Wu J, Zhang X, Guo X, Wang W. 2014. Comparative analysis of mitochondrial genomes in distinct nuclear ploidy loach Misgurnus anguillicaudatus and its implications for polyploidy evolution. PLOS One. 9(3):e92033.