Abstract
In the context of Indian zoogeography, the DNA barcode data of short-horned grasshoppers (family Acrididae) are limited in global databases. Hence, the present study was aimed to collect selected Acridid species from the Indian Himalayan regions and generate DNA barcode data to enrich the global database. The estimated K2P genetic distances, Bayesian analysis (BA) topology and multiple species delimitation methods (ABGD, bPTP, and GMYC) clearly discriminate all the studied species. Based on high genetic distance (7.5%), multiple clades, and more than one molecular operational taxonomic unit, the present study elucidates the allopatric speciation and presence of possible cryptic diversity of Oxya japonica within India, China, and Russia. The present study suggests the collection of multiple specimens from different geographical locations and the generation of more DNA barcode data would facilitate the actual diversity of this insect group.
Introduction
Orthoptera is one of the oldest living insect lineages with 28,418 described species and contributes almost half of the total arthropods diversity globally (Grimaldi and Engel Citation2005; Cigliano et al. Citation2020). They are generally inhabits in the grassland ecosystems and play an important role in maintaining the food chain (Belovsky and Slade Citation1993; Joern et al. Citation2006). Due to their high mobility across different geographical locations and ecological sensibilities, this group of insects also acts as a bio-indicator (Samways and Sergeev Citation1997; Bazelet and Samways Citation2014). Among the extant diversity, most of the species were considered as major pests in agriculture, animal husbandry, and forest ecosystem (Uvarov Citation1966; COPR Citation1982). In recent past, the United Nations’ Food and Agriculture Organization (FAO) declared that a single Acridid species has made upsurge invasion and worst outbreak as well as damaged several crops in India and neighboring countries. However, few of them were used as a bio-control agent; e.g. the semi-aquatic grasshopper species, Cornops aquaticum has been used as a bio-control agent on aquatic plant (Bownes et al. Citation2013) and the meadow purple-striped grasshopper, Hesperotettix viridis feed the noxious snakeweeds which can harm cattle and other livestock (Thompson and Richman Citation1993). Besides its ecological and economic significance, the family Acrididae also has a biogeographic history and assumed to have originated in the early Cenozoic Era and diversified through mid to late Cenozoic (Song et al. Citation2015).
The short-horned grasshopper (family Acrididae) is the largest and most diverse lineage within the suborder Caelifera and comprised around 6700 species within 26 subfamilies worldwide (Cigliano et al. Citation2020). So far, India shares 2.4% of the global orthopteran diversity, and the family Acrididae is represented by 285 species under 135 genera (Shishodia et al. Citation2010). In the last 100 years, the uncommon specimens of orthopterans confronted taxonomic challenges or cover-splitting on the systematics concepts, which often forcefully sorted them as Acrididae members (Eades Citation2000; Song Citation2010). Nevertheless, the species identification of Acridid orthopterans is often challenging due to high phenotypic plasticity (Simpson and Sword Citation2009), polymorphism (Ichikawa et al. Citation2006), and lack of identification keys for sub-adults (Bellmann Citation2006). Molecular tools such as DNA barcoding have been largely supplemented with systematics studies for addressing species identification and other biological questions (Hebert et al. Citation2003; Tyagi et al. Citation2017). This advanced technology has also evidenced to discriminate different taxonomic groups including orthopterans (Huang et al. Citation2013; Singha et al. Citation2019; Tyagi et al. Citation2019). As of now, several studies have been conducted to elucidate the accurate species identification, estimation of genetic divergence, phylogenetic relationship, detection of cryptic diversity, and species complexes of this insect group (Wang et al. Citation2008; Chapco and Contreras Citation2011; Song et al. Citation2018). However, majority of the studies have been focused on three subfamilies, Melanoplinae (Chapco et al. Citation2001; Amédégnato et al. Citation2003), Oedipodinae (Chapco et al. Citation1997; Fries et al. Citation2007), and Gomphocerinae (Bugrov et al. Citation2005; Chapco and Contreras Citation2011). Nevertheless, the generation of DNA barcode data of several Acrididae species is still underway from different geographical regions especially from India, which restrict the assumption on their actual diversity. Hence, the present study was attempted to generate 20 partial mitochondrial cytochrome c oxidase subunit I (mtCOI) sequences of six morphologically identified Acridid species from the Indian Himalayan regions (IHRs). We used genetic distance matrix, Bayesian analysis (BA), and multiple species delimitation methods to elucidate their genetic diversity. This is the first and preliminary attempt which advocated further large-scale effort to illuminate the extant orthopteran diversity from the IHRs.
Materials and methods
Total 20 short-horned grasshoppers specimens were sampled from five different locations (27.49N 96.39E, 27.54N 96.44E, 27.53N 91.67E, 27.71N 91.73E, 27.53N 91.72E) in Arunachal Pradesh state in the eastern Himalayan region (). The expedition was conducted under the National Mission on Himalayan Studies project entitled ‘Biodiversity Assessment through Long-term Monitoring Plots in Indian Himalayan Landscape’ project ID: NMHS/2015-16/LG-05; project grant number: NMHS/LG2016/0011/8509 at Zoological Survey of India (ZSI) (Kolkata, India). The hind leg of each specimen was dissected through surgical blade and preserved in 70% ethanol at −20 °C for further molecular analysis. The specimens were dried, pinned, preserved, and identified up to species level by reviewing the available literatures (Bolívar Citation1914; Kirby Citation1914; Ramme Citation1940; Hollis Citation1971; Jago Citation1984; Hollis Citation2009; Swaminathan et al. Citation2018) (Table S1). Nikon digital camera (D-7000) was used to acquire the photograph of the representative species. The voucher specimens were stored in the National Zoological Collections of the Orthoptera section, ZSI (Kolkata, India).
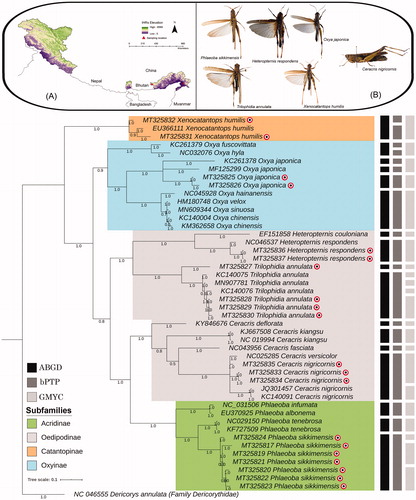
Figure 1. (A) The collection locality map of the studied Acrididae species in IHRs. (B) The vouchered specimens of the morphologically identified six Acrididae species. (C) The Bayesian analysis showed distinct clades of all the studied Acrididae species with high posterior probability support. The generated sequences are marked by color dots. Gray shades color bars beside the tree indicate the delineated MOTUs by different species delimitation methods (ABGD, bPTP, and GMYC).
Total genomic DNA was extracted from each morphologically identified specimen in 500 µl of tissue lysis buffer containing 50 mM Tris–HCl, 25 mM EDTA, 150 mM NaCl, and proteinase K (200 µg/ml) through phenol–chloroform extraction method (Sambrook and Russell Citation2006). To generated the barcode data, degenerative primer pair: COBU (5′-TYTCAACAAAYCAYAARGATATTGG-3′) and COBL (5′-TAAACTTCWGGRTGWCCAAARAATCA-3′) was used to amplify the partial fragment of mitochondrial cytochrome c oxidase subunit I (COI) gene (Pan et al. Citation2006). The amplification was performed in 30 μl reaction mixture of 100 ng of DNA template, 1× PCR buffer, 2 mM MgCl2, 10 pmol of each primer, 0.25 mM of each dNTPs, 0.25 U of high-fidelity Taq DNA polymerase (Applied Biosystems Inc., Foster City, CA), and nuclease free water. The amplification was carried out in VeritiVR Thermal Cycler (Applied Biosystems, Foster City, CA) with the following thermal profile: an initial denaturation step at 95 C for 5 min, followed by 30–35 cycles of denaturation at 94 C for 45 s, annealing at 48 C for 45 s, and extension at 70 C for 1 min 30 s, and a final extension at 72 C for 10 min and then held at 4 C. The PCR products were purified by QIAquickR Gel extraction kit (Qiagen Inc., Germantown, MD) with manufactures protocol. The cycle-sequencing was performed by using BigDye®Terminator ver. 3.1 Cycle Sequencing Kit (Applied Biosystems, Inc., Foster City, CA) and 3.2 pmol of each primer on the same thermal cycler. The cycle-sequencing products were purified by using BigDye X-terminator kit (Applied Biosystems Inc., Foster City, CA, USA) and sequenced bi-directionally through 48 capillary ABI 3730 Genetic Analyzer housed at ZSI (Kolkata, India).
Both forward and reverse contigs of each sample were screened by Sequence Scanner (Applied Biosystems, Foster City, CA, USA) to generate the consensus sequences. Each sequence was examined through the nucleotide BLAST program (https://blast.ncbi.nlm.nih.gov) and ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) to diminish the indels, mismatches, and start-stop codons. The generated sequences were submitted in the GenBank database through Bankit submission tool (https://www.ncbi.nlm.nih.gov/WebSub/?tool=genbank) to acquire the accession numbers. Further, to make an analytical dataset, total 26 barcode sequences of same or related species under four targeted subfamilies were downloaded from GenBank database. The barcode sequence of Dericorys annulata: accession no. NC_046555 (Family: Dericorythidae) was also acquired from GenBank and used as an out-group in the present topology analysis to test the monophyletic criterion. The final dataset were aligned using ClustalX program (Thompson et al. Citation1997) and the genetic divergences were estimated by Kimura 2 parameter (K2P) in MEGA7 (Kumar et al. Citation2016). The best fit model for all three codon positions ‘GTR + I + G’ was estimated by observing the lowest BIC value through PartitionFinder version 2.1.1 (Lanfear et al. Citation2012). To check the monophyletic clustering of the studied taxa, BA was followed by using Mr Bayes 3.1.2 (Ronquist and Huelsenbeck Citation2003). The dataset was run for 50,000,000 generations with 25% burn-in with trees saving at every 100 generation. The Markov Chain Monte Carlo (MCMC) analysis was used to generate the convergence metrics, till the standard deviation of split frequencies reached 0.01 and the potential scale reduction factor for all parameters bordered on 1.0. To visualize the topology with better representation, the tree was modified through web based iTOL tool (https://itol.embl.de/) (Letunic and Bork Citation2007). In addition to understand the below species-level diversity, three species delimitation methods: Automatic Barcode Gap Discovery (ABGD) (Puillandre et al. Citation2012), the General Mixed Yule-coalescent (GMYC) (Fujisawa and Barraclough Citation2013), and Poisson-Tree-Processes (bPTP) (Zhang et al. Citation2013) were executed to estimate the molecular operational taxonomic units (MOTUs). The ABGD analysis was carried out on the web interface (www.abi.snv.jussieu.fr/public/abgd/) with Simple, K80 Kimura, and JC69 Jukes-Cantor Distance with relative gap width (X = 1.5). The ultrametric tree was generated by BEAST program (Drummond and Rambaut Citation2007 ) with GTR + I + G model and the Tree Annotator was used for visualizing the output tree. For GMYC analysis, multiple threshold was carried out in RStudio (https://www.r-project.org/) using packages like ‘ape’ and ‘splits’ (Paradis et al. Citation2004; Ezard et al. Citation2009). The maximum-likelihood tree was further built in RAxML (Stamatakis Citation2006) to execute the bPTP analysis on web interface (http://species.h-its.org/ptp/).
Results and discussion
Many large-scale attempts have been made to generate the DNA barcode data of orthopterans from diverse geographical regions around the world. However, a single attempt has been made from India focusing on the family Tettigoniidae so far (Muhammedali et al. Citation2017). This restricts the knowledge on the actual diversity of orthopterans from the Indian biogeography, especially from the IHRs. The present study intended to enrich the global barcode library of the orthopteran insect through an integrated approach from the IHRs. The 20 studied samples were identified as six morphospecies under four Acrididae subfamilies (Acridinae, Catantopinae, Oedipodinae, Oxyinae) (). Among the generated DNA barcodes, seven sequences of Phlaeoba sikkimensis are novel contributions to the global database. To confirm the molecular identification of the studied taxa and monophyletic criteria, the genetic divergence and topological clustering were tested for the final dataset (46 DNA barcode sequences of 20 species under four Acridid subfamilies). The overall mean genetic distance of the studied dataset was 13.9%. Excluding the singleton taxa, the mean intra-species genetic distance was ranging from 0% (Ceracris kiangsu, Phlaeoba tenebrosa) to 7.5% (Oxya japonica).
The highest inter-species genetic distance (20.8%) was depicted between Xenocatantops humilis and Heteropternis couloniana. Further, the significantly low genetic distances were observed in few Oxyinae species: Oxya chinensis and Oxya velox (0.3%), Oxya sinuosa and O. velox (0.2%), as well as O. sinuosa and O. chinensis (0.5%). The low genetic distance was also detected for two Acridinae species, Phlaeoba infumata and Phlaeoba albonema (0.5%). Due to the low sample coverage and use of single gene analysis, we suggest multiple specimens and additional molecular information is required to validate the systematics status of these species. The present BA analysis showed distinct clades for all the studied species with high posterior probability support which is congruent with the earlier cladistics hypothesis (Song et al. Citation2018). The multiple species delimitation methods: ABGD, GMYC, and bPTP yielded 17 (Table S2), 19 (Table S3), and 22 (Table S4) MOTUs, respectively ().
Moreover, the previous studies suggested that >3% intra-species genetic distances can be used as a threshold for delimiting arthropod species (Hebert et al. Citation2003; Huang et al. Citation2013). However, this barcode gap may vary among different taxa due to the unlike molecular evolutionary rate within the mitochondrial genes (Wiemers and Fiedler Citation2007; Meier et al. Citation2008). Adding to this, a recent molecular study also implied different species delimitation methods for resolving cryptic species and species complex among different Orthopteran families (Zhou et al. Citation2019). However, the present study detected considerable high intra-species genetic distance (7.5%) and more than one MOTU in O. japonica, which indicates the presence of possible cryptic diversity (). This Oxyinae species is commonly known as rice grasshopper and originally described from Japan. The species act as a pest on rice and widely distributed throughout Asia, Africa, Northern Africa, and Algeria (Cigliano et al. Citation2020). A total of two specimens of O. japonica were collected from Tawang, Arunachal Pradesh in the eastern Himalayan region. The studied specimens of O. japonica along with the available database sequences (KC261378 collected from Russia and MF125299 collected from China) showed polytomy in BA tree and more than one MOTU in ABGD, bPTP, and GMYC analyses. By superimposing all species delimitation methods, the present molecular-based investigation presumed the presence of cryptic diversity of O. japonica in India, China, and Russia. Further, the collection localities of O. japonica from distant geographical areas suggest the occurrence of allopatric speciation of this species within its range-distribution. In conclusion, the present study contributed first DNA barcode data of morphologically identified selected Acridid species from the IHRs. We encourage more rigorous sampling of this taxonomic group from different geographical regions and generation of more barcode data to elucidate their actual species diversity.
Supplemental Material
Download MS Word (17.4 KB)Acknowledgements
The authors are thankful to the Director, Zoological Survey of India (ZSI), Ministry of Environment, Forests and Climate Change (MoEF&CC), Govt. of India for providing necessary facilities, constant support, and encouragement throughout the study. The second author (SK) acknowledges the fellowship grant received from the Council of Scientific and Industrial Research (CSIR) Senior Research Associateship (Scientists’ Pool Scheme) Pool No. 9072-A. We thank Devkant Singha and Avas Pakrashi for analytical help and Pronomay Karmakar for helping in improving the language.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank database at https://www.ncbi.nlm.nih.gov with the accession numbers MT325817, MT325819–MT325837 which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Additional information
Funding
References
- Amédégnato C, Chapco W, Litzenberger G. 2003. Out of South America? Additional evidence for a southern origin of melanopline grasshoppers. Mol Phylogenet Evol. 29(1):115–119.
- Bazelet CS, Samways MJ. 2014. Habitat quality of grassland fragments affects dispersal ability of a mobile grasshopper, Ornithacris cyanea (Orthoptera: Acrididae). Afr Entomol. 22(4):714–725.
- Bellmann H. 2006. Der Kosmos-Heuschreckenf€uhrer. Stuttgart: Kosmos.
- Belovsky GE, Slade JB. 1993. The role of vertebrate and invertebrate predators in a grasshopper community. Oikos. 68(2):193–201.
- Bolívar I. 1914. Estudios entomológicos. Segunda parte. I. El grupo de los Euprepocnemes. II. Los Truxalinos del antiguo mundo. Trabajos del Museo de Ciencias Naturales (Serie Zoológica). 20:110.
- Bownes A, Hill MP, Byrne MJ. 2013. Nutrient-mediated effects on Cornops aquaticum Bruner (Orthoptera: Acrididae), a potential biological control agent of water hyacinth, Eichhornia crassipes (Mart.) Solms (Pontederiaceae). Biol Control. 67(3):548–554.
- Bugrov A, Novikova O, Mayorov V, Adkison L, Blinov A. 2005. Molecular phylogeny of Palaearctic genera of Gomphocerinae grasshoppers (Orthoptera, Acrididae). Syst Entomol. 31(2):362–368.
- Chapco W, Contreras D. 2011. Subfamilies Acridinae, Gomphocerinae and Oedipodinae are “fuzzy sets”: a proposal for a common African origin. J Orthoptera Res. 20(2):173–190.
- Chapco W, Litzenberger G, Kuperus WR. 2001. A molecular biogeographic analysis of the relationship between North American melanoploid grasshoppers and their Eurasian and South American relatives. Mol Phylogenet Evol. 18(3):460–466.
- Chapco W, Martel RKB, Kuperus WR. 1997. Molecular phylogeny of North American band-winged grasshoppers (Orthoptera: Acrididae). Ann Entomol Soc Am. 90(5):555–562.
- Cigliano MM, Braun H, Eades DC, Otte D. 2020. Orthoptera species file. Version 5.0/5.0 [11.X.2019]. http://Orthoptera.SpeciesFile.org
- COPR (Centre for Overseas Pest Research). 1982. The locust and grasshopper agricultural manual. London (UK): Centre for Overseas Pest Research.
- Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 7:214.
- Eades DC. 2000. Evolutionary relationships of phallic structures of Acridomorpha (Orthoptera). J Orthoptera Res. 9(9):181–210.
- Ezard T, Fujisawa T, Barraclough TG. 2009. Splits: species’ limits by threshold statistics. R package version 1.0-14/r31. http://R-Forge.R-project.org/projects/splits/
- Fries M, Chapco W, Contreras D. 2007. A molecular phylogenetic analysis of the Oedipodinae and their intercontinental relationships. J Orthoptera Res. 16(2):115–125.
- Fujisawa T, Barraclough TG. 2013. Delimiting species using single-locus data and the generalized mixed Yule coalescent approach: a revised method and evaluation on simulated data sets. Syst Biol. 62(5):707–724.
- Grimaldi D, Engel MS. 2005. Evolution of the insects. New York: Cambridge University Press; p. 755.
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc Biol Sci. 270(1512):313–322.
- Hollis D. 2009. A revision of the genus Trilophidia Stal (Orthoptera: Acridoidea). Trans R Entomol Soc Lond. 117(8):245–262.
- Hollis D. 1971. A preliminary revision of the genus Oxya Audinet-Serville (Orthoptera: Acridoidea). Bull Br Mus Nat Hist Entomol. 26:269–343.
- Huang J, Zhang A, Mao S, Huang Y. 2013. DNA barcoding and species boundary delimitation of selected species of Chinese Acridoidea (Orthoptera: Caelifera). PLOS One. 8(12):e82400.
- Ichikawa A, Kano Y, Kawai M, Tominago O, Murai T. 2006. Orthoptera of the Japanese Archipelago in Color. Hokkaido: Hokkaido University Press; p. 688.
- Jago ND. 1984. The alate genera of east African Catantopinae (Orthoptera: Acridoidea) including revision of the genus Catantops Schaum. Trans Am Entomol Soc. 110:295–387.
- Joern A, Danner BJ, Logan JD, Wolesensky W. 2006. Natural history of mass-action in predator–prey models: a case study from wolf spiders and grasshoppers. Am Midl Nat. 156(1):52–62.
- Kirby WF. 1914. Fauna of British India, including Ceylon and Burma. Orthoptera (Acrididae); 276 pp.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lanfear R, Calcott B, Simon Y, Ho W, Stephane GS. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701.
- Letunic I, Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23(1):127–128.
- Meier R, Zhang G, Ali F. 2008. The use of mean instead of smallest interspecific distances exaggerates the size of the “barcoding gap” and leads to misidentification. Syst Biol. 57(5):809–813.
- Muhammedali VC, Akhilesh VP, Sebastian CD. 2017. DNA barcoding for identification of Conocephalus dorsalis (Orthoptera: Tettigoniidae) from Northern Kerala using cytochrome oxidase subunit I gene. Int Res J Biol Sci. 6:8–10.
- Pan CY, Hu J, Zhang X, Huang Y. 2006. The DNA barcoding application of mtDNA COI gene in seven species of Catantopidae (Orthoptera). Entomotaxonomia. 28:103–110.
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 20(2):289–290.
- Puillandre N, Lambert A, Brouillet S, Achaz G. 2012. ABGD, automatic barcode gap discovery for primary species delimitation. Mol Ecol. 21(8):1864–1877.
- Ramme W. 1940. Beiträge zur Kenntnis der Acrididen-Fauna des indomalayischen und benachbarter Gebiete (Orth.). Mit besonderer Berücksichtigung der Tiergeographie von Celebes. Mitt. Mus. Nat.kd. Berl., Zool. Reihe. 25(1):1–243.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.
- Sambrook J, Russell DW. 2006. Purification of nucleic acids by extraction with phenol: chloroform. Cold Spring Harb Protoc. 2006(1):169–170.
- Samways MJ, Sergeev MG. 1997. Orthoptera and landscape change. In: Gangwere SK, Muralirangan MC, Muralirangan M, editors. The bionomics of grasshoppers, katydids and their kin. Wallingford (UK): CAB International; p. 147–162.
- Shishodia MS, Chandra K, Gupta SK. 2010. Records of the Zoological Survey of India. In Director, editor. An annotated checklist of Orthoptera (Insecta) from India. Kolkata, India: Zoological Survey of India; p. 1–366.
- Simpson SJ, Sword GA. 2009. Phase Polyphenism in Locusts: Mechanisms, Population, Consequences, Adaptive Significance and Evolution. In: Whitman DW and Ananthakrishnan TN editors. Phenotypic Plasticity of Insects: Mechanisms and Consequences. Science Publishers Inc., Plymouth. p. 147-190.
- Singha D, Kumar V, Chakraborty R, Kundu S, Hosamani AK, Kumar V, Tyagi K. 2019. Molecular footprint of Frankliniella occidentalis from India: a vector of Tospoviruses. Mitochondr DNA B Resour. 4(1):39–40.
- Song H. 2010. Grasshopper systematics: past, present and future. J Orthoptera Res. 19(1):57–68.
- Song H, Amédégnato C, Cigliano MM, Desutter-Grandcolas L, Heads SW, Huang Y, Otte D, Whiting MF. 2015. 300 million years of diversification: elucidating the patterns of orthopteran evolution based on comprehensive taxon and gene sampling. Cladistics. 31(6):621–651.
- Song H, Mariño-Pérez R, Woller DA, Cigliano MM. 2018. Evolution, diversification, and biogeography of grasshoppers (Orthoptera: Acrididae). Insect Syst Divers. 2:1–25.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Swaminathan R, Nagar R, Swaminathan T. 2018. Representative species of the tribe Catantopini (Orthoptera: Acrididae) from India. Trans Am Entomol Soc. 144(2):239–261.
- Thompson DC, Richman DB. 1993. A grasshopper that only eats snakeweed? In: Sterling TM, Thompson DC, editors. Research Report 674, Agricultural Experiment Station Cooperative Extension Service New Mexico State University, Snakeweed research updates and highlights. New Mexico Agricultural Experimental Station Research Report; p. 18–19.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25(24):4876–4882.
- Tyagi K, Kumar V, Kundu S, Pakrashi A, Prasad P, Caleb JTD, Chandra K. 2019. Identification of Indian spiders through DNA barcoding: cryptic species and species complex. Sci Rep. 9(1):14033.
- Tyagi K, Kumar V, Singha D, Chandra K, Laskar BA, Kundu S, Chakraborty R, Chatterjee S. 2017. DNA barcoding studies on thrips in India: cryptic species and species complexes. Sci Rep. 7(1):4898.
- Uvarov BP. 1966. Grasshoppers and locusts. Vol. 1. Cambridge (UK): Cambridge University Press.
- Wang NX, Feng X, Jiang GF, Fang N, Xuan WJ. 2008. Molecular phylogenetic analysis of five subfamilies of the Acrididae (Orthoptera: Acridoidea) based on the mitochondrial cytochrome b and cytochrome c oxidase subunit I gene sequences. Acta Entomol Sin. 51:1187–1195.
- Wiemers M, Fiedler K. 2007. Does the DNA barcoding gap exist? A case study in blue butterflies (Lepidoptera: Lycaenidae). Front Zool. 4:8.
- Zhang J, Kapli P, Pavlidis P, Stamatakis AA. 2013. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 29(22):2869–2876.
- Zhou Z, Guo H, Han L, Chai J, Che X, Shi F. 2019. Singleton molecular species delimitation based on COI-5P barcode sequences revealed high cryptic/undescribed diversity for Chinese katydids (Orthoptera: Tettigoniidae). BMC Evol Biol. 19(1):79.
