Abstract
Paphiopedilum spicerianum (H. G. Reichenbach) Pfitzer is an endangered and threatened orchid species in China. The genetic and molecular data of this orchid species is deficient. With the aim to identify appropriate chloroplast markers for the use in a conservation biology study, the complete chloroplast genome of P. spicerianum was reported. We found that the chloroplast genome of P. spicerianum is 157,292 bp in length, containing a large single-copy region of 87,252 bp, a small single-copy region of 1828 bp, and a pair of inverted repeats of 34,106 bp. Genome annotation predicted 105 unique genes, including 71 protein-coding genes, 30 tRNAs, and four rRNAs. Fourteen genes contained one intron and two genes (clpP and ycf3) had two introns. The GC content of the P. spicerianum was 35.8%. Phylogenetic analysis indicated P. spicerianum was closely related to P. purpuratum.
Paphiopedilum spicerianum (H. G. Reichenbach) Pfitzer is an endangered orchid with significant ornamental values. It is restricted to eastern and northeastern Myanmar, Thailand, and southwestern China. In China, it is known from only one site in Yunnan Province (Ye and Luo Citation2006). Due to habitat loss and limited populations, P. spicerianum is listed as one of the wild plants with extremely small population by State Forestry Administration of China (Ma et al. Citation2013). With the popularity of Next-Seq technology, coupled with the moderate length and evolution rate of chloroplast genome, chloroplast genome has been gradually used for species evolution analysis, and can also be used for the establishment of conservative strategies for endangered plant.
The fresh leaves of P. spicerianum were collected from orchid nursery of Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Yunnan Province, China (21°54′N, 101°46′E; voucher specimen: LY0154, deposited in Xishuangbanna Tropical Botanical Garden Herbarium, HITBC). Total genomic DNA from leaf was extracted by using Tiangen DNA kit (TIANGEN, Beijing, China), and sequenced by the Illumina Hiseq 2500 sequencing platform (Illumina, Foster City, CA) at Personal Biotechnology Co., Ltd. (Shanghai, China). The total 3.0 G data of high-quality reads were trimmed, and then used to assemble the chloroplast genome using a toolkit named GetOrganelle (Jin et al. Citation2018). The assembled chloroplast genome was annotated by the web server GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html, Tillich et al. Citation2017). The tRNA genes were verified by tRNAscan-SE v2.0.3 (Lowe and Chan Citation2016). The annotated complete chloroplast genome of P. spicerianum was deposited in GenBank with accession number MT683624.
The complete chloroplast genome of P. spicerianum was 1,57,292 bp in length, which presented a typical quadripartite structure, and consists of a large single-copy region (LSC, 87,252 bp), a small single-copy region (SSC, 1828 bp), and two inverted repeat regions (IRs, 34,106 bp). The total GC content of chloroplast genome was 35.8%, whereas the corresponding values of LSC, SSC, and IR regions are 33.2%, 23.8%, and 39.5%, respectively. Paphiopedilum spicerianum chloroplast genome encoded 105 unique genes, including 71 protein-coding genes, 30 tRNA genes, and four rRNA genes. Fourteen genes had a single intron while the clpP and ycf3 genes had two introns.
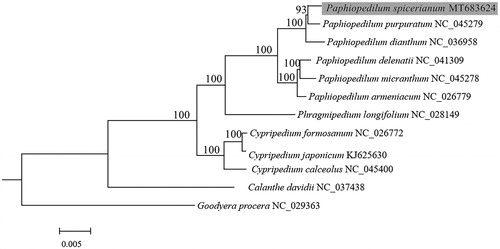
To further investigate its phylogenetic position, the complete chloroplast genomes from 10 species of Cypripedioideae, and one species from Orchidoideae and one species from Epidendroideae as outgroup, were selected to reconstruct phylogenetic tree. Sixty-five protein-coding genes exported from Geneious Primer 2020 (Biomatters, Auckland, New Zealand) were aligned with MAFFT (Katoh and Standley Citation2013) and Mauve (Darling et al. Citation2004). A maximum-likelihood (ML) analysis of the plastome data and Best-fit model TVM + F+I + G4 were performed using IQ-TREE-2.0.5 with 1000 bootstrap replicates (Minh et al. Citation2020). Phylogenetic analysis showed that P. spicerianum was closely related to P. purpuratum within Cypripedioideae with strong support, and the taxa of Paphiopedilum formed a well-supported clade with Phragmipedium longifolium ().
Figure 1. Phylogenetic position of Paphiopedilum spicerianum (gray box) inferred by the maximum-likelihood (ML) analysis based on 65 protein-coding genes from chloroplast genome using 1000 bootstrap replicates. The number at each node indicates bootstrap value and the scale is substitution per site.
Acknowledgements
We are grateful Fei Zhao in Kunming Institute of Botany, Chinese Academy of Sciences, Yu Song, Wen Bin Yu, from Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, for their help in genomic data analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genomic sequence data that support the findings of this study have been deposited in GenBank with accession no. MT683624 (https://www.ncbi.nlm.nih.gov/).
Additional information
Funding
References
- Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14(7):1394–1403.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57.
- Ma YP, Chen G, Grumbine ED, Dao ZL, Sun WB, Guo HJ. 2013. Conserving plant species with extremely small populations (PSESP) in China. Biodivers Conserv. 22(3):803–809.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler AV, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Ye DP, Luo YB. 2006. Paphiopedilum spicerianum: a new record of Orchidaceae from China. Acta Phytotax Sin. 44(4):471–473.
