Abstract
Colored rice is gaining popularity due to its use in creative agriculture and the value of healthy consumption. However, the quality and yield characteristics of most rice varieties still need to be improved. Revealing the genetic background of colored rice is of great significance to promote crop improvement. Here, the completed chloroplast (cp) genome sequence of yellow colored Oryza sativa voucher HSAGSDYD1802 was sequenced and reported. It was a 134,502 bp circular DNA with a typical quadripartite structure, consisting of two reverse repeat regions (IRa and IRb, 20,804 bp) separated by a large single-copy region (LSC, 80,547 bp) and a small single-copy region (SSC, 12,347 bp). The total GC content was 39%. The cp genome encoded 146 genes, containing 100 protein-coding genes, 8 rRNAs, and 38 tRNA genes. Phylogenetic analysis indicated that O. sativa voucher HSAGSDYD1802 was closely related to O. sativa L. TN1, RP Bio-226 and IR8. This study enriches the genetic information of colored rice and is helpful for future molecular breeding.
Rice field art is a large-scale artwork that uses different leaf-color Oryza sativa varieties to draw pictures or letters in the paddy field. In recent years, rice field art has been popular in China and other countries (Kim et al. Citation2015). In addition, colored rice is rich in phenolic antioxidants, which has important health benefits and production value for the supply of healthy rice. So far, colored rices, including red, purple and yellow varieties, have been grown in Asia (Patel et al. Citation2014). However, most varieties suffer from low grain yield, leading most farmers to be less interested in growing colored rice. Hybrid breeding techniques can improve the yield of colored rice, the success of crop improvement also depends on the genetic characteristics of hybrid rice parents. Therefore, the lack of understanding of the genetic background of hybrid parents will be disadvantageous to rice breeding.
Compared with the nuclear genome, chloroplast genome shows uni-parental inheritance, high conserved genomic structure, low nucleotide recombination and mutation rates, and is widely used to determine the evolutionary relationship of plants at different taxonomic levels (Fan et al. Citation2019). The chloroplast genome sequences have been characterized in some conventional rices (Yu et al. Citation2017). However, the chloroplast genome sequences of colored rice have been rarely reported. Here, we described the chloroplast genome sequence of O. sativa voucher HSAGSDYD1802, a yellow ornamental rice widely used in creative agriculture.
Rice plants were grown in the rice base of Leshan Normal University, Leshan, Sichuan Province, China (103°44′57″E, 29°33′53″N). The specimen (No. HSAGSDYD1802) was kept in the molecular laboratory of Leshan Normal University. Genomic DNA was isolated from fresh rice leaves, an Illumina paired-end library with a size of 300 bp inserts was constructed and sequenced using the Illumina HiSeqXten platform. Subsequently, the quality control and assembly of sequencing readings were carried out by NGS QC tool Kit v2.3.3 and SPAdes v.3.11.0 software, respectively (Bankevich et al. Citation2012; Patel and Jain Citation2012). Finally, the complete chloroplast genome sequence was annotated using the PGA software (Qu et al. Citation2019) and a circular genome map was drawn using the OGDRAW program (Lohse et al. Citation2007).
The chloroplast genome of O. sativa voucher HSAGSDYD1802 had a quadripartite structure with a length of 134,502 bp (GenBank accession no. MT653617). In which, the size of large single-copy (LSC) and small single-copy region (SSC) were 80,547 bp and 12,347 bp, respectively, the length of the two inverted repeats (IRs) was 20,804bp. It contained 146 genes (121 unique genes), including 100 protein-coding genes (87 unique genes), 8 rRNAs (4 unique genes) and 38 tRNA genes (30 unique genes), most of which were single-copy genes. However, 12 protein-coding genes (ndhB, ORF103, ORF107, ORF131, ORF255, ORF99, rpl2, rpl23, rps15, rps19, rps7, ycf76), 4 rRNA genes (rrn16, rrn23, rrn4.5, rrn5) and 8 tRNA genes (trnA-UGC, trnH-GUG, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC) were repeated in the IR regions, the rps12 gene was trans-splicing. In addition, the results showed that 8 protein-coding genes (rps16, atpF, petB, petD, rpl16, rpl2, ndhB, ndhA) and 6 tRNA genes (trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnA-UGC, trnI-GAU) contained one intron, ycf3 gene contained two introns. The total GC content of chloroplast genome was 39%.
To confirm the phylogeny of O. sativa voucher HSAGSDYD1802, its complete chloroplast genome sequence was compared with that of other thirty-two O. sativa cultivars using MAFFT7.037 software (Katoh and Standley Citation2013), and then the Maximum-Likelihood (ML) phylogenetic tree was constructed by Mega-X v10.0.5 software (Kumar et al. Citation2018). The program operating parameters were set as follows: a Tamura 3-parameter (T92) nucleotide substitution model with 1000 bootstrap repetitions, accompanied by Gamma distributed with Invariant site (G + I) rates, and partial deletion of gaps/missing data. The result of ML phylogenetic tree showed that O. sativa voucher HSAGSDYD1802 was closely related to O. sativa cultivar TN1, RP Bio-226 and IR8 ().
Figure 1. Phylogeny of the complete chloroplast genome sequence of O. sativa voucher HSAGSDYD1802. Note: The maximum-likelihood (ML) phylogenetic tree was constructed based on 33 chloroplast genome sequences. Numbers near the branch indicated the probability obtained by 1,000 bootstrap analysis, probability values less than 50% were not shown.
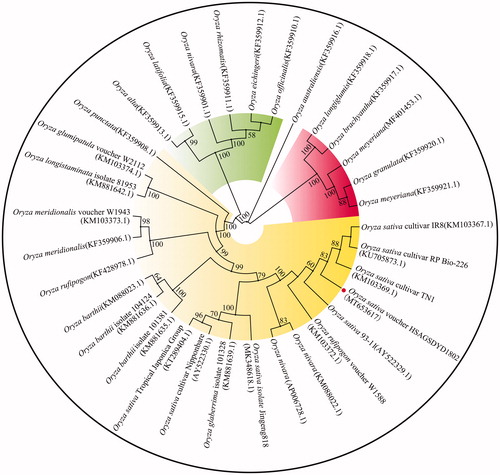
Due to the low yield of colored rices, most materials have not yet been subject to variety certification, resulting in unclear cytoplasmic genetic information for many materials. Chloroplast genome sequences have been widely used in rice germplasm and kinship identification (Waters et al. Citation2012). In this study, the chloroplast genome sequence of O. sativa voucher HSAGSDYD1802 was obtained by the Illumina HiSeqXten platform sequencing. Sequence comparison showed that O. sativa voucher HSAGSDYD1802 was clustered into the indica rice group, but had unique nucleotide sites. In summary, this study provides a useful genomic resource for molecular identification and phylogenetic study of colored rice. In addition, the chloroplast genome data will also help to develop plastid genetic markers.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data supporting the results of this study is available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number [MT653617], or available from the corresponding author.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Fan J, Fu QC, Liang Z. 2019. Complete chloroplast genome sequence and phylogenetic analysis of Sinojackia sarcocarpa, an endemic plant in Southwest China. Mitochondr DNA Part B. 4(1):1350–1351.
- Katoh K, Standley DM. 2013. MAFFT Multiple sequence alignment software Version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim CK, Seol YJ, Shin Y, Lim HM, Lee GS, Kim AR, Lee TH, Lee JH, Park DS, Yoo S, et al. 2015. Whole-genome resequencing and transcriptomic analysis to identify genes involved in leaf-color diversity in ornamental rice plants. PLOS One. 10(4):e0124071.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619.
- Patel S, Ravikiran R, Chakraborty S, Macwana S, Sasidharan N, Trivedi R, Aher B. 2014. Genetic diversity analysis of colored and white rice genotypes using microsatellite (SSR) and Insertion-Deletion (INDEL) markers. Emir J Food Agric. 26(6):497–507.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Waters DLE, Nock CJ, Ishikawa R, Rice N, Henry RJ. 2012. Chloroplast genome sequence confirms distinctness of Australian and Asian wild rice. Ecol Evol. 2(1):211–217.
- Yu Y, Lee HO, Chin JH, Park HY, Yoo SC. 2017. The complete chloroplast genome sequence of Oryza sativa aus-type variety Nagina-22 (Poaceae). Mitochondr DNA Part B. 2(2):819–820.
