Abstract
The Asiatic softshell turtle, also known as the black-rayed softshell turtle (Amyda cartilaginea; Accession no: MT039230), is found in northeastern India (Mizoram), Brunei Darussalam, Indonesia, Malaysia, Singapore, Myanmar, Laos, Vietnam, Cambodia, and Thailand. This turtle is thought to have been introduced into the Sunda Islands, Sulawesi, and Yunnan, China, through the Malay Peninsula to Sumatra, Java, and Borneo. Herein, we determined the complete mitochondrial genome of A. cartilaginea for the first time using next-generation sequencing (NGS). The assembled mitogenome was 16,763 bp in length and encoded 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA genes (12S rRNA and 16S rRNA), and one control region (CR). The PCGs based maximum-likelihood phylogeny discriminated A. cartilaginea from other Testudines and clusters within family Trionychidae with the sister taxa of Nilssonia nigricans.
Asiatic softshell turtle or black-rayed softshell turtle (Amyda cartilaginea), is widely distributed in northeastern India (Mizoram), Brunei Darussalam, Indonesia, Malaysia, Singapore, Myanmar, Laos, Vietnam, Cambodia, and Thailand. Besides, the turtle is thought to have been introduced into the Sunda Islands, Sulawesi, and Yunnan, China, through the Malay Peninsula to Sumatra, Java, and Borneo (Iverson Citation1992; Kuchling Citation1995; IUCN Citation2000; Koch et al. Citation2008; Van Dijk et al. Citation2012; Fritz et al. Citation2014; Kundu et al. Citation2016). Like other softshell turtles, A. cartilaginea exhibits a leathery shell skin, a pig-nose-like snout, four webbed feet for underwater swimming, and three powerful claws on each foot for killing and dissecting prey (Fritz et al. Citation2014). As a relatively large-sized freshwater turtle, the shell of an adult A. cartilaginea usually exceeds 40 cm in length (Van Dijk Citation1992) and can grow up to 83 cm (Ernst et al. Citation2000). Asiatic softshell turtle is listed as a vulnerable (VU) species by the International Union for Conservation of Nature (IUCN Citation2000), following years of overexploitation as pets and bush meat(Kundu et al. Citation2016). Softshell turtles vary in color and body proportions, as such morphological data is of limited use in taxonomy and systematics. The application of modern molecular techniques has improved the understanding of the taxonomy of softshell turtles, and thus focus is increasingly being directed in the generation and analysis of molecular genetics data (Fritz et al. Citation2014). This study aimed to determine the complete mitogenome of A. cartilaginea and its phylogenetic relationships with other turtle species. The findings of the present study will lay a foundation for future research and protection of A. cartilaginea.
We extracted muscle tissue samples from dead Asiatic softshell turtles found at the Dianchi Lake, Kunming, Yunnan, China. The samples were stored at the Museum of Kunming Institute of Zoology, Yunnan province, China (Institute coordinates: 25°4′12.87″N, 102°42′12.05″E., elev. 1900 m). Total genomic DNA was extracted from the muscle tissues using the Ezup pillar genomic DNA extraction kit (Sangon Biotech, Shanghai, China). Next-generation sequencing (NGS) of the mitochondrial genome was performed by the Personal Biotechnology Co, Ltd (Shanghai, China), as described previously (Metzker Citation2010). Briefly, we first constructed approximately 400 bp insert size libraries using a Whole Genome Shotgun (WGS) strategy. The libraries were sequenced using a 2 × 250 bp paired-end (PE) protocol based on the Illumina MiSeq sequencing platform. Next, the 38,596,792 reads were filtered in the raw data to generate a high quality (HQ) clean data using Adapter Removal v2 and SOAPec v2.01software. The filtered 37,868,126 reads were then used for de novo assembly by A5-miseq v20150522 and SPAdes v3.9.0 software. BLASTn (BLASTC v2.2.31+) in NCBI was applied to identify the contigs of mitogenome sequences. Pilon v1.18 was used to evaluate the accuracy and completeness of the genome assembly. Finally, the complete mitogenome sequence with validated annotations was submitted to GenBank with accession number MT039230.
The mitogenome of A. cartilaginea was to be 16,763 bp in length and encodes 37 genes, including 13 protein-coding genes (PCGs), two ribosomal RNA (12S rRNA, 16S rRNA), 22 transfer RNA genes (tRNAs), and one mitochondrial control region (CR or D-loop). The base composition is 36.2% A, 25.7% T, 26.3% C, and 11.8% G, demonstrating a bias of higher AT content (61.9%) than GC content (38.1%). The positions of all genes were predicted by the MITOS Web Server (Bernt et al. Citation2013). All the tRNA genes are encoded on the H-strand, except for tRNAGln, tRNAAla, tRNAAsn, tRNACys, TrnaTyr, tRANSer(UCN), TrnaGlu, and TrnaPro genes. All the PCGs are encoded on the H-strand except nad6, which is the only PCG encoded on the L-strand. Most of the PCGs begin with an ATG initiation codon; only COI starts with GTG. Besides, 11 PCGs end with complete TAA, TAG, or AGA termination codon, whereas the remaining two COIII and ND4 are terminated by incomplete TA(A) or T(AA) termination codon, respectively. Furthermore, we found an additional nucleotide that causes a translational frameshift in nad3. Like most turtles hitherto studied, the gene distribution of A. cartilaginea was identical to the typical vertebrate mitochondrial genome (Boore Citation1999).
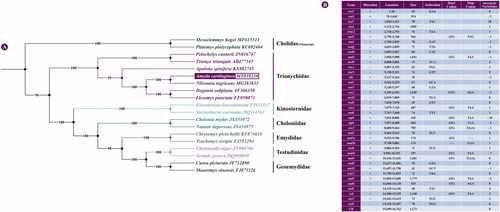
All 13 PCGs datasets were first concatenated using PhyloSuite v1.2.2 (Zhang et al. Citation2020). The concatenated alignment consisted of 11,322 bp for 19 species from seven families (Chelidae, Trionychidae, Kinosternidae, Cheloniidae, Emydidae, Testudinidae, and Geoemydidae). Two side-necked turtles (family: Chelidae) were selected as outgroup. Maximum likelihood (ML) phylogenies of A. cartilaginea were inferred using IQ-TREE v1.6.8 (Nguyen et al. Citation2015) under the Edge-linked partition model for 5000 ultrafast bootstraps (Minh et al. Citation2013). The phylogenetic trees based on the mitogenomes indicated that A. cartilaginea has the closest relationship with N. nigricans and clusters within the clade of family Trionychidae (). These findings may shed light on future investigations into the molecular evolution and conservation of softshell turtle species.
Figure 1. Phylogenetic tree and genomic annotation of A. cartilaginea. (A) Maximum-likelihood (ML) phylogenetic tree yielded by IQTREE for seven turtle families based on a concatenated alignment of 13 protein-coding genes from 19 turtle species; bootstrap values are shown in the middle of the branches connected to the nodes. (B) Annotation of the complete mitogenome of A. cartilaginea.
Acknowledgments
Thanks to Mengjiao Zhou for her help in language editing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data used to support the findings of this study are available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/MT039230, reference number MT039230.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Ernst C, Altenburg R, Barbour R. 2000. Turtles of the world. World biodiversity database, version 1.2 [CD-ROM]. Amsterdam: Biodiversity Center of ETI.
- Fritz U, Gemel R, Kehlmaier C, Vamberger M, Praschag P. 2014. Phylogeography of the Asian softshell turtle Amyda cartilaginea (Boddaert, 1770): evidence for a species complex. Vertebr Zool. 64(2):229–243.
- IUCN. 2000. Amyda cartilaginea (errata version published in 2016). The IUCN Red List of Threatened Species 2000: e.T1181A97397687. Downloaded on 07 August 2020; [accessed]. https://dx.doi.org/10.2305/IUCN.UK.2000.RLTS.T1181A3309466.en.
- Iverson JB. 1992. A revised checklist with distribution maps of the turtles of the world. Homestead (FL): Green Nature Books.
- Koch A, Ives I, Arida E, Iskandar D. 2008. On the occurrence of the Asiatic softshell turtle, Amyda cartilaginea (Boddaert, 1770), on Sulawesi, Indonesia. Hamadryad. 33:121–127.
- Kuchling G. 1995. Turtles at a market in western Yunnan: possible range extensions for some southern Asiatic chelonians in China and Myanmar. Chelonian Conserv Biol. 1:223–226.
- Kundu S, Kumar V, Laskar BA, Chandra K, Tyagi K. 2016. Mitochondrial DNA effectively detects non-native Testudines: invisible wildlife trade in northeast India. Gene Rep. 4:10–15.
- Metzker ML. 2010. Sequencing technologies – the next generation. Nat Rev Genet. 11(1):31–46.
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Van Dijk P. 1992. Variation in the Southeast Asian soft-shelled turtle, Amyda cartilaginea [unpublished MSc thesis]. Galway: National University of Ireland, 79:13.
- Van Dijk P, Iverson J, Shaffer H, Bour R, Rhodin A. 2012. Turtles of the world, 2012 update: annotated checklist of taxonomy, synonymy, distribution, and conservation status. Chelonian Res Monogr. 5:000.243–000.328.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
