Abstract
The mason bee, Osmia pedicornis Cockerell, 1919, which is importantly used as the pollinator, particularly for apples in Korea. We sequenced the mitochondrial genome (mitogenome) of O. pedicornis as an initial study for species identification and subsequent population genetic study. The size of the incomplete genome was 14,505 bp, excluding the trnA, trnC, and the A + T-rich region that were unable to sequence, but including partially sequenced trnM and srRNA. The genome included typical sets of protein-coding genes (PCGs), rRNA genes, and one non-coding region, tRNAs, excluding two unidentified tRNAs. Although positions of the two tRNAs that were not sequenced are unknown the gene arrangement of O. pedicornis mitogenome has the tRNA arrangement, trnM-trnQ-trnI, at the A + T-rich region and ND2 junction that differed from that of previously published O. excavate, which has trnA-trnQ-trnI arrangement at the junction. Phylogenetic analyses were performed using concatenated sequences of the 13 PCGs genes and the maximum likelihood method with the inclusion of a total of 12 mitogenome sequences belonging to three families in the superfamily Apoidea. Current O. pedicornis was placed as the sister to the O. bicornis, with the highest nodal support. The Apidae and Megachilidae were placed as the sister group, with the placement of Colletidae as the basal lineage for the group with the highest nodal support.
Bees are one of the most effective pollinators accounting for 16,325 species in the world (Michener Citation2007). The mason bee Osmia Panzer, 1806 (Hymenopetra: Megachilidae) includes 339 species (Michener Citation2007). The mason bee differs from the honey bee in that the individuals are solitary, all females are fertile, make her own nest, and no worker bees exist (Torchio Citation1989; Bosch and Kemp Citation2001; Lee et al. Citation2016). Osmia pedicornis Cockerell, 1919 is distributed in Korea, eastern China, and Japan (Yasumatsu and Hirashima Citation1950) and is importantly used as the pollinator, particularly for apples (Cane Citation2008; Yoon et al. Citation2016).
In Korea, three species of Osmia (O. cornifrons, O. pedicornis, and O. taurus) are occurring, but no mitochondrial genome (mitogenome) sequences are available. Therefore, we sequenced O. pedicornis mitogenome as an initial study for species identification, selection of variable regions for subsequent population genetic study, and phylogenetic reconstruction of the genus and higher taxonomic groups in Apoidea to which Osmia is included.
In 2016, one adult of O. pedicornis was collected in Muneundan-ro, Nam-myeon, Jeongseon-gun, Gangwon-do, Republic of Korea (37°16′03′′ N, 128°44′29′′ E) and subsequently deposited at the Chonnam National University, Korea, under accession no. CNU12885. DNA was extracted from the hind legs of this specimen using a Wizard Genomic DNA Purification Kit (Promega, Madison, WI). Three long overlapping fragments (LFs: COI-ND4, ND5-lrRNA, and lrRNA-COI) were amplified from the genomic DNA and 28 short overlapping fragments were subsequently amplified using the LFs as templates. The primers for LFs and SFs were designed using two available Osmia mitogenomes (Zheng et al. Citation2018; Unpublished, GenBank acc. nos. KT164643, KT164653, and KT164669). A direct sequencing by Sanger’s method after PCR amplification was performed for majority of SFs, but where impossible sequencing was performed after cloning. Phylogenetic analysis was performed using 12 available mitogenomes from the clade Anthophila in the superfamily Apoidea including the one obtained in this study (). Nucleotide sequences of 13 protein-coding genes (PCGs) were aligned and concatenated (10,993 bp excluding gaps). An optimal partitioning scheme (eight partitions) were determined using PartitionFinder 2 and the Greedy algorithm (Lanfear et al. Citation2012, Citation2014, Citation2016). Maximum likelihood (ML) analysis was performed using RAxML-HPC2 on XSEDE version 8.0.24 (Stamatakis Citation2014), implemented on the CIPRES Portal version 3.1 (Miller et al. Citation2010).
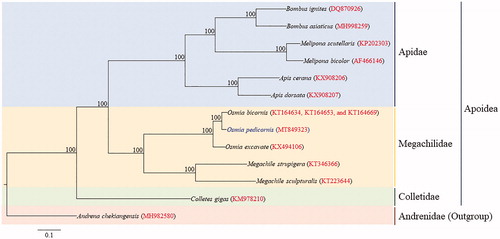
Figure 1. Maximum likelihood (ML) method-based phylogenetic tree for the superfamily Apoidea using concatenated sequences of 13 protein-coding genes. The numbers at each node specify the bootstrap percentages of 1000 pseudoreplicates. The scale bar indicates the number of substitutions per site. Andrena chekiangensis belonging to the family Andrenidae in the superfamily Apoidea (MH982580, He et al. Citation2019) was utilized as an outgroup. The GenBank accession numbers are as follows: Bombus asiaticus, MH998259 (Zhao et al. Citation2019); B. ignitus, DQ870926 (Cha et al. Citation2007); Apis cerana, KX908206 (Wang et al. Citation2018); A. dorsata, KX908207 (Wang et al. Citation2018); Melipona bicolor, AF466146 (Silvestre et al. Citation2008); M. scutellans, KP202303 (Pereira et al. Citation2016); Osmia pedicornis, MT849323 (This study); O. excavata, KX494106 (Zheng et al. Citation2018); O. bicornis, KT164634, KT164653, and KT164669 (Unpublished); Megachile sculpturalis, KT223644 (Zhang et al. Citation2017); M. strupigera, KT346366 (Huang, Su, He, et al. Citation2016); and Colletes gigas, KM978210 (Huang, Su, Qu, et al. Citation2016).
Although a substantial attempt to sequence whole mitogenome of the O. pedicornis was made we eventually were unable to sequence a serial genes located around the A + T-rich region, such as trnA, trnC, trnM (partially), and srRNA (partially) including the A + T-rich region. Probably this may happen due to the unexpectedly long A + T-rich region, which possibly contains the repeat sequences and higher A/T nucleotide, along with nonspecific amplification. In fact, the O. excavata A + T-rich region expands to 1472 bp, but is still incomplete and contains several repeat sequences, ranging in size from 9 to 38 bp (Zheng et al. Citation2018).
The size of the O. excavata mitogenome was 14,505 bp, excluding the trnA, trnC, and the A + T-rich region that were unable to sequence, but including partially sequenced trnM and srRNA. The size and A/T content of the O. pedicornis PCGs was 3684 codons (excluding termination codons) and 83.9%, respectively, and are similar to that of O. excavata (3682 codons and 82.4%, respectively). The size and A/T content of O. pedicornis lrRNA was 1328 bp and 86.2% and also is similar to those of O. excavata (1320 bp and 85.8%, respectively).
The gene arrangement of O. excavata mitogenome, which lacks for the information for two tRNA positions (trnA and trnC) differed from that of O. excavata mitogenome (Zheng et al. Citation2018) in that the five tRNA region located at the A + T-rich region and ND2 junction has an trnM-trnQ-trnI arrangement, instead of the trnA-trnQ-trnI arrangement in O. excavate (Zheng et al. Citation2018).
Phylogenetic analysis using nucleotide sequences of 13 PCGs with the representative mitogenome sequences of Apoidea showed that each Apidae and Megachilidae, which were represented by multiple sequences formed monophyletic groups with the highest nodal supports (). Among three families represented for Apoidea the Apidae and Megachilidae were placed as the sister group, leaving Colletidae as the basal lineage for the group with the highest nodal support. Within Megachilidae, to which Osmia is included three species of Osmia formed a monophyletic group with the highest nodal support, forming O. pedicornis and O. bicornis the sister group. This Osmia group was placed as the sister to two Megachile species. A recent whole genome sequence-based phylogenetic analysis including several families of Apoidea has also shown the sister relationship between Apidae and Megachilidae with the placement of Colletidae as the basal lineage for the group (Zhou et al. Citation2020), presenting an identical phylogenetic relationship to current results ().
Disclosure statement
No potential conflicts of interest are reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccore/MT849323.1
Additional information
Funding
References
- Bosch J, Kemp WP. 2001. How to manage the blue orchard bee, Osmia lignaria, as an orchard pollinator. Washington (DC): Sustainable Agriculture Network.
- Cane JH. 2008. Bees (Hymenoptera: Apoidea: Apiformes). Encyclo Entomol. 2:419–434.
- Cha SY, Yoon HJ, Lee EM, Yoon MH, Hwang JS, Jin BR, Han YS, Kim I. 2007. The complete nucleotide sequence and gene organization of the mitochondrial genome of the bumblebee, Bombus ignitus (Hymenoptera: Apidae). Gene. 392(1–2):206–220.
- He B, Su T, Niu Z, Zhou Z, Gu Z, Huang D. 2019. Characterization of mitochondrial genomes of three Andrena bees (Apoidea: Andrenidae) and insights into the phylogenetics. Int J Biol Macromol. 127:118–125.
- Huang D, Su T, He B, Gu P, Liang AP, Zhu C. 2016. Sequencing and characterization of the Megachile strupigera (Hymenoptera: Megachilidae) mitochondrial genome. Mitochondrial DNA Part B. 1(1):282–284.
- Huang D, Su T, Qu L, Wu Y, Gu P, He B, Xu X, Zhu C. 2016. The complete mitochondrial genome of the Colletes gigas (Hymenoptera: Colletidae: Colletinae). Mitochondrial DNA Part A. 27(6):3878–3879.
- Lee KY, Yoon HY, Lee KS, Jin BR. 2016. Development and mating behavior of Osmia cornifrons (Hymenotpera: Megachilidae) in the constant temperature. J Asia Pac Entomol. 19(2):281–287.
- Michener CD. 2007. The bees of the world. Baltimore (MD): Johns Hopkins University Press.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the 9th Gateway Computing Environments Workshop (GCE), New Orleans (LA): p. 1–8.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701.
- Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A. 2014. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol Biol. 14:82.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. Partition finder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol E. 34:772–773.
- Pereira UDP, Bonetti AM, Goulart LR, Santos ARD, Oliveira GCD, Cuadros-Orellana S, Ueira-Vieira C. 2016. Complete mitochondrial genome sequence of Melipona scutellaris, a Brazilian stingless bee. Mitochondrial DNA Part A. 27(5):3387–3388.
- Silvestre D, Dowton M, Arias MC. 2008. The mitochondrial genome of the stingless bee Melipona bicolor (Hymenoptera, Apidae, Meliponini): sequence, gene organization and a unique tRNA translocation event conserved across the tribe Meliponini. Genet Mol Biol. 31(2):451–460.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Torchio PF. 1989. Biology, immature development, and adaptive behavior of Stelis montana, a cleptoparasite of Osmia spp. (Hymenoptera: Megachilidae). Ann Entomol Soc Am. 82(5):616–632.
- Wang AR, Kim JS, Kim MJ, Kim HK, Choi YS, Kim I. 2018. Comparative description of mitochondrial genomes of the honey bee Apis (Hymenoptera: Apidae): four new genome sequences and Apis phylogeny using whole genomes and individual genes. J Apic Res. 57(4):484–503.
- Yasumatsu K, Hirashima Y. 1950. Revision of the genus Osmia of Japan and Korea hymenoptera: megachilidae. Mushi. 21:1–18.
- Yoon HJ, Lee KY, Kim SY, Kim YM, Kwon CR. 2016. Distribution and ecological characteristics of cocoons of the solitary Bees Osmia cornifrons and O. pedicornis (Hymenoptera: Megachilidae). Apiculture. 31(3):183–194.
- Zhang Y, Su T, He B, Gu P, Huang D, Zhu C. 2017. Sequencing and characterization of the Megachile sculpturalis (Hymenoptera: Megachilidae) mitochondrial genome. Mitochondrial DNA Part A. 28(3):344–346.
- Zhao F, Yan J, Jiang K, Huang Z, Lin G. 2019. Nearly complete mitochondrial genomes of four bumblebee species (Hymenoptera: Apidae: Bombus). Mitochondrial DNA Part B. 4(1):183–184.
- Zheng BY, Cao LJ, Tang P, van Achterberg K, Hoffmann AA, Chen HY, Chen XX, Wei SJ. 2018. Gene arrangement and sequence of mitochondrial genomes yield insights into the phylogeny and evolution of bees and sphecid wasps (Hymenoptera: Apoidea). Mol Phylogenet E. 124:1–9.
- Zhou QS, Luo A, Zhang F, Niu ZQ, Wu QT, Xiong M, Orr MC, Zhu CD. 2020. The first draft genome of the plasterer bee Colletes gigas (Hymenoptera: Colletidae: Colletes). Genome Biol Evol. 12(6):860–866.
