Abstract
The bamboo aphids, Pseudoregma spp., are the commonest insect pests found in ornamental bamboos throughout southeastern Asia. In this study, the mitochondrial genome of a representative of Pseudoregma bambucicola isolated from the bamboo Bambusa multiplexcv in Guizhou of China was determined through Illumina MiSeq platform. The entire genome was 16,705 bp in length and encoded 13 protein-coding genes, 22 tRNA genes, and 2 rRNA genes. The phylogeneric analysis showed that the P. bambucicola (Guizhou isolate) clustered together with another two isolates from Sichuan and Fujian of China, respectively, and together formed a monophyletic relationship with Hormaphis betulae in Hormaphidinae. The mitochondrial DNA data presented here should contribute to future molecular identification, population genetic, and evolutionary biological studies of P. bambucicola.
The aphids of the genus Pseudoregma are the common insect pests of bamboos that are widely distributed throughout the warmer regions of southeastern Asia (Fukatsu et al. Citation2001). These aphid species, for instance, Pseudoregma alexanderi, P. bambucicola, P. Carolinensis, and P. koshunensis mainly infest ornamental bamboos and cause their growth stunting and even cause death (Fukatsu et al. Citation2001; Ijichi et al. Citation2004; Nong et al. Citation2017). Until now there has been significant advance of knowledge made in morphology, ecologics, behavior and chemical control as well as systematics of Pseudoregma spp., especially in P. bambucicola (Fukatsu et al. Citation2001; Nong et al. Citation2017, Citation2019a, Citation2019b; Zhang et al. Citation2019). Nevertheless, it is still existing gaps in the understanding P. bambucicola including its molecular epidemiology and population genetic diversity due to limited marker resources (Nong et al. Citation2017, Citation2019a, Citation2019b; Zhang et al. Citation2019). Mitochondrial (mt) DNA is a valuable marker resource and is being widely used for genetics and molecular identification of plant aphids (Cameron Citation2014; De Mandal et al., Citation2014; Marquina et al. Citation2019). In this study, we characterized the complete mitochondrial genome of a representative of P. bambucicola sampled from Guizhou of China and added novel mt DNA data to the this species.
In May 2020, about 200 aphids were sampled from the Bambusa multiplexcv which was planted in Guiyang city (27°07′N, 107°05′E), Guizhou province, China. These aphid specimens were identified as P. bambucicola according to the taxonomic key of Stern (Citation1997) and the molecular sequencing by amplification of the mt cox2 and cytb genes (Nong et al. Citation2019a, Citation2019b). Twenty aphid specimens were pooled for mt DNA extraction and the remaining were fixed in 5% formalin solution and archived in the Insect Museum of Bamboo Diseases and Pest control and Resources Development Key Laboratory of Sichuan Province, China, under voucher number NX2019_14. The mt genome was sequenced using the Illumina MiSeq platform (Novogene, Beijing, China). The genome was assembled by MITObim (Hahn et al. Citation2013) and annotated using MITOS (Bernt et al. Citation2013). The complete genome sequence has been deposited in GenBank under accession number: MT916291.
The mitochondrial genome of P. bambucicola (Guizhou isolate) was 16,705 bp in length and encoded 13 protein-coding genes (PCGs), 22 tRNA genes (tRNAs), and 2 rRNA genes (rRNAs). Similar to the congeneric species (Zhang et al. Citation2019; Nong et al. Citation2020), nine PCGs and 15 tRNAs were found to be transcribed on the forward strand (J-strand) while the remaining genes were located on the reverse strand (N-strand). Across the 13 PCGs, except for the nad4 deduced to use an incomplete stop codon ‘T’, the rest were predicted to use the typical TAG (n = 5) or TAA (n = 7) as the stop codon. Twenty-two tRNAs ranged from 51 bp (tRNA-Cys) to 73 bp (tRNA-Lys) in length and all can be folded into typical clover-leaf like secondary structures, with the exception of tRNA(AGN)-Ser. Within two rRNAs, the large (rrnL; 1,284 bp) and small rRNA (rrnS; 758 bp) subunits were located between tRNA(CUN)-Leu and tRNA-Val and between tRNA-Val and D-loop region, respectively. The D-loop region (849 bp) with 95.9% A + T content was present between rrnS and tRNA-Ile. In addition, a total of 207 bp intergenic spacers were present at 17 positions and the lengths of the spacers were 1–27 bp.
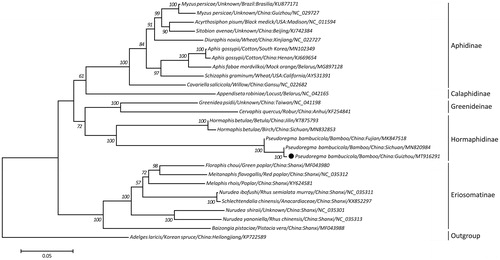
Based on a concatenated amino acid sequence of 13 protein-coding genes from 27 aphids, a maximum-likelihood (ML) phylogeny was reconstructed using Adelges laricis as the outgroup. The phylogeneric analysis showed that P. bambucicola (Guizhou isolate) clustered with two isolates from Sichuan and Fujian of China, respectively, and together formed a monophyletic relationship with Hormaphis betulae in the subfamily Hormaphidinae, with 100% bootstrapping confidences (). In addition, within this tree topology, Aphidinae, Calaphidinae, Greenideinae, Eriosomatinae, and Hormaphidinae were treated as monophyletic groups, consistent with results of recent molecular studies (Wang et al. Citation2013; Li Citation2017; Zhang et al. Citation2019), demonstrating the phylogenetic stability of these subfamily in Aphididae. Taken together, the complete mt genome of P. bambucicola (Guizhou isolate) characterized here should contribute to a better understanding of phylogenetic relationships among Aphididae species and also serve molecular identification, population genetic and evolutionary biological studies of P. bambucicola.
Figure 1. A maximum likelihood (ML) tree inferred from concatenated amino-acid sequences of 13 mitochondrial protein-coding genes of P. bambucicola and other related aphids, utilizing MtArt model with 1,000,000 bootstrap replications (<50% support not shown). The solid black cycle represents the species in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT916291.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Ann Rev Entomol. 59:95–117.
- De Mandal S, Chhakchhuak L, Gurusubramanian G, Kumar NS. 2014. Mitochondrial markers for identification and phylogenetic studies in insects–a review. DNA Barcodes. 2(1):1–9.
- Fukatsu T, Shibao H, Nikoh N, Aoki S. 2001. Genetically distinct populations in an Asian soldier-producing aphid, Pseudoregma bambucicola (Homoptera: Aphididae), identified by DNA fingerprinting and molecular phylogenetic analysis. Mol Phylogenet Evol. 18(3):423–433.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Ijichi N, Shibao H, Miura T, Matsumoto T, Fukatsu T. 2004. Soldier differentiation during embryogenesis of a social aphid, Pseudoregma bambucicola. Entomol Sci. 7(2):143–155.
- Li YQ, Chen J, Qiao GX. 2017. Complete mitochondrial genome of the aphid Hormaphis betulae (Mordvilko) (Hemiptera: Aphididae: Hormaphidinae). Mitochondrial DNA Part B. 28(2):265–266.
- Marquina D, Andersson AF, Ronquist F. 2019. New mitochondrial primers for metabarcoding of insects, designed and evaluated using in silico methods. Mol Ecol Resour. 19(1):90–104.
- Nong X, Wang L, Liu Y, Zhong S, Yu X, Xie Y. 2020. The complete mitochondrial genome of the bamboo aphid Pseudoregma bambucicola and its phylogenetic position. Mitochondrial DNA Part B. 5(1):642–643.
- Nong X, Zeng X, Yang Y, Liang Z, Tang M, Liao L, Luo C. 2017. Morphological observation and characterization of the Pseudoregma bambucicola with the scanning electron microscope. Saudi J Biol Sci. 24(7):1626–1630.
- Nong X, Zhong SN, Chen L, Yang YJ, Xie Y, Luo CB, Fu C, Yu H, Liang Z. 2019a. Genetic differentiation of populations of Pseudoregma bambucicola based on mtDNA cytb gene sequences. Mitochondrial DNA Part B. 4(1):1803–1807.
- Nong X, Zhong SN, Li SM, Yang YJ, Liang Z, Xie Y. 2019b. Genetic differentiation of Pseudoregma bambucicola population based on mtDNA COII gene. Saudi J Biol Sci. 26(5):1032–1036.
- Stern DL, Whitfield JA, Foster WA. 1997. Behavior and morphology of monomorphic soldiers from the aphid genus Pseudoregma (Cerataphidini, Hormaphididae): implications for the evolution of morphological castes in social aphids. Insectes Soc. 44(4):379–392.
- Wang Y, Huang XL, Qiao GX. 2013. Comparative analysis of mitochondrial genomes of five aphid species (Hemiptera: Aphididae) and phylogenetic implications. PLoS One. 8(10):e77511.
- Zhang H, Deng J, Liu Q, Huang X. 2019. The mitochondrial genome of a social aphid, Pseudoregma bambucicola (Hemiptera: Aphididae: Hormaphidinae). Mitochondrial DNA B Resour. 4(2):2100–2101.
